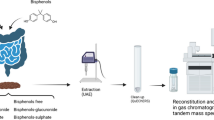
Abstract
The widespread occurrence of antibiotics in the environment may exert a negative impact on wild organisms. In addition, they can become environmental reservoirs, through the ingestion of food or contaminated water, and vectors for antibiotic-resistant bacteria. This fact is even more important in migratory birds that can promote their dissemination across continents. In this work, a multiresidue analytical method suitable for the determination of five families of antibiotics and their main metabolites in waterbird faeces has been developed and validated. The target compounds include environmentally significant sulfonamides, macrolides, fluoroquinolones, tetracyclines and antifolates. Sample treatment involves ultrasound-assisted extraction with methanol and dispersive solid-phase extraction clean-up with C18. Analytical determination was carried out by liquid chromatography–tandem mass spectrometry. The most significant parameters affecting sample extraction and extract clean-up were optimised by means of experimental designs. Good linearity (R2 > 0.994), accuracy (from 41 to 127%), precision (relative standard deviation lower than 24%) and limits of quantification (lower than 2 ng g−1 (dry weight, dw)) were obtained for most of the compounds. The method was applied to the determination of the selected compounds in 27 faeces samples from three common migratory waterbird species. Nine antibiotics and three of their metabolites were detected in the analysed samples. Fluoroquinolones and macrolides were the antibiotics most frequently detected. The highest concentrations corresponded to norfloxacin (up to 199 ng g−1 dw).
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The widespread use of antibiotics in human and veterinary medicine may lead to their environmental dissemination through discharges from wastewater treatment plants (WWTPs), animal farms and aquaculture ponds [1]. The presence of residues of fluoroquinolones, sulfonamides, tetracyclines or macrolides, among other antibiotics, has been reported in aquatic and terrestrial environmental compartments at part per billion or part per million levels [2,3,4]. In spite that their environmental concentrations are significantly lower than their therapeutic use concentrations, there is increasing evidence that such concentrations may cause antibiotic resistance which implies not only a threat to human and animal health but also a food safety risk [5,6,7]. This situation is even more worrying in the case of migratory birds because they can promote the international transfer of bacterial antibiotic resistance [8]. Migratory birds are considered a major source of the spread of antibiotic resistance across different environments such as water supplies and landfills as well as over long distances across habitats and regions [9,10,11]. Recently, Jarma et al. [9] found different bacterial communities in faeces from waterbirds wintering in Spain. Their results suggest that these birds may disseminate antibiotic resistance between landfills and natural habitats even in pristine environments such as Antarctica [12].
To assess the presence of antibiotics in birds, some methods have been proposed for their determination in feathers [13,14,15,16,17,18,19,20]. However, biomonitoring in feathers has several drawbacks such as being an invasive matrix that, foremost, requires bird capture which is costly and not always feasible. Moreover, such studies are limited to the determination of parent compounds not including the determination of metabolites. Metabolites of antibiotics can be present in wild birds not only because of metabolic transformations of the parent compounds but also by direct exposition via diet because of their presence in the aquatic and terrestrial environments. Therefore, analytical methods to evaluate the exposition of migratory birds to antibiotics should include not only parent compounds but also their metabolites. In addition, a non-invasive matrix such as faeces would be useful to evaluate risk assessment of wild birds, to calculate environmental loads and to evaluate environmental dissemination of antibiotic residues [9]. Analytical methods reported to date for the determination of pharmaceuticals in animal excrements have been focused on bovine manure [21,22,23], swine manure [21,22,23,24,25,26,27,28] and poultry excreta [2, 29, 30]. The aim of such studies is to evaluate the presence of veterinary antibiotics in manure or excreta not only because they can constitute a significant route of antibiotic dissemination into the environment but also because they could enter in the human food chain when manure is used in crops as low-cost organic fertilisers. Because of that, as can be seen above, most of the studies have been conducted in excrements from mammals, and much less from (domestic) birds. Nevertheless, analytical methods developed for their application to mammals’ excreta are not suitable for birds’ excreta because of the differences between both types of excreta. Whereas mammals excrete nitrogenous wastes mostly in the form of urea, birds convert it to uric acid or guanine, which is eliminated simultaneously with faeces through the cloaca (unlike mammals, which have separate mechanisms to eliminate faeces and urine) [31]. In addition, in spite that some analytical methods have been reported for the determination of pharmaceuticals in excreta from domestic birds, it can differ in their physical–chemical composition (based on the diet, flight activity, metabolism, physiological conditions, caecum microbiome, manipulated diet, etc.) from excreta from wild birds. For example, magnesium is commonly dietarily manipulated in domestic chickens to modify the consistency (moisture) of excreta [32]. Because of all the mentioned above, it is necessary to develop analytical methods for the specific determination of antibiotics and their metabolites in wild bird faeces as they constitute promising to non-invasive and easy to collect matrix to monitor exposure to antibiotics.
Therefore, the aim of this work was to optimise and validate an analytical method for the determination of twelve environmentally relevant antibiotics from five different families, and eight of their main metabolites, in wild bird faeces samples. Target compounds included three sulfonamides, four macrolides, three fluoroquinolones, a tetracycline, an antifolate and eight of their metabolites. The selection of the antibiotics was based on their presence in the environment [2,3,4]. Only one tetracycline was included in the analytical method because, in spite of being wide-spectrum antibiotics widely used in human and veterinary medicine, they are present at low levels in the aquatic environment as they precipitate with cations being retained onto wastewater treatment plants’ sewage sludge and sediments [2]. Sample treatment was based on easy-to-perform and low-cost techniques: ultrasonic-assisted extraction (UAE) and extract clean-up by dispersive solid-phase extraction (d-SPE). The method was applied to bird faeces from three different migratory waterbird species wintering in Doñana National Space and surrounding areas (Andalusia, South Spain). The application allowed us to evaluate the exposure of birds and their role as biovectors of antibiotic residuals, and to correlate these data with the diversity and abundance of the antibiotic resistance gene.
Materials and methods
Chemicals and reagents
Antibiotic standards of enrofloxacin (ENR, ≥ 98.5%), ciprofloxacin (CIP, ≥ 98.0%), sulfadiazine (SDZ, ≥ 99.0%), N4-acetylsulfadiazine (AcSDZ, > 99.0%), N4-acetylsulfamethoxazole (AcSMX, ≥ 98.5%), sulfamethazine (SMZ, ≥ 99.0%), roxithromycin (ROX, ≥ 98.0%) and tetracycline (TC, ≥ 95.0%) were purchased from Sigma-Aldrich (Steinheim, Germany). Sulfamethoxazole N4-glucoside (SMX-GL, > 99.0%), N4-acetylsulfamethazine (AcSMZ, ≥ 98.0%), N-desmethylclarithromycin (DM-CLM, ≥ 96.0%), 4-hydroxytrimethoprim (4-OH-TMP, ≥ 97.0%) and 3-desmethyltrimethoprim (DM-TMP, ≥ 98.0%) were supplied by Toronto Research Chemicals (Toronto, Canada). Clarithromycin (CLM, ≥ 98.0%) and erythromycin (ERY, ≥ 98.0%) were purchased from Tokyo Chemicals Industry (Eschborn, Germany). Trimethoprim (TMP, ≥ 99.5%), sulfamethoxazole (SMX, ≥ 99.5%) and norfloxacin (NOR, ≥ 99.1%) were supplied by Dr Ehrenstorfer GmbH (Augsburg, Germany). 4-Epitetracycline (EP-TC, > 99.0%) was provided from WHO Centre for Chemical Reference Substances (Strasbourg, France). Azithromycin (AZM, > 99.0%) was supplied by European Pharmacopoeia Reference Standard (Strasbourg, France). The internal standards (I.S.) ofloxacin-d3 (OFL-d3, ≥ 99.0%), demeclocycline (DMC, ≥ 90.0%), sulfamethoxazole-(phenyl-13C6) (SMX-13C, ≥ 99.0%) and erythromycin-(N,N-dimethyl-13C2) (ERY-13C, ≥ 99.0%) were purchased from Sigma-Aldrich (Steinheim, Germany). The structures of the target compounds, and their pKa and log Kow values, are presented in Table S1 in Electronic Supplementary Material (ESM).
Ammonium formate and Florisil® were supplied by Sigma-Aldrich (Madrid, Spain). Primary-secondary amine (PSA), ammonium acetate, C18 and glacial acetic acid were provided by Scharlab (Barcelona, Spain). Formic acid was provided by Panreac (Barcelona, Spain). The reagents were of high analytical grade and purity. LC–MS-grade acetonitrile (ACN), hexane, acetone, water and methanol (MeOH) were supplied by Biosolve BV (Valkenswaard, the Netherlands).
Sample collection and treatment
Faeces samples corresponded to three migratory waterbird species: white stork (Ciconia ciconia), lesser black-backed gull (Larus fuscus) and black-headed gull (Chroicocephalus ridibundus). Freshly voided faeces were collected from roosting habitats in Doñana National Space (Andalusia, Spain). Monospecific flocks were previously located with binoculars. Then, we approached what caused birds to fly away, and then fresh faeces, easily distinguished from older ones, were collected from the core of the faecal material by using a spatula. To avoid collecting faeces from the same bird, samples were collected at least 2 m one from the other. They were transported to the laboratory in a cooler bag with a cooling block and then they were conserved at − 20 °C until analysis.
After collection, samples were freeze-dried in a Cryodos-50 lyophiliser (Telstar, Terrasa, Spain), homogenised in a mortar and sieved (particle size < 100 µm). Pre-treated faeces samples (0.5 g dry weight (dw)) were spiked with the I.S. (OFL-d3, DMC, SMX-13C and ERY-13C) at 100 ng g−1 dw. Samples were extracted three times with 5 mL of MeOH by sonication in an ultrasonic bath (25 °C, 80 kHz) for 10 min. After each extraction, the solid–liquid separation was carried out by centrifugation for 10 min at 2900 × g. The liquid phases were combined into a clean centrifuge tube containing 0.8 g of C18 for d-SPE clean-up. The tubes were shaken and centrifuged for 10 min at 2900 × g. The liquid phase was transferred to another tube to be evaporated to dryness under a gentle nitrogen stream. The extract was reconstituted in 0.5 mL of MeOH:water (1:1, v/v) and filtered through a 0.22 µm cellulose syringe filter. A 2 µL aliquot of the filtered extract was injected into the liquid chromatography–tandem mass spectrometry (LC–MS/MS) instrument.
Instrumental analysis
Chromatographic determination was performed using an Agilent 1260 Infinity II chromatograph (Agilent, Santa Clara, CA, USA). Chromatographic separation was carried out in a Zorbax RRHD Eclipse Plus C18 (150 mm × 3.0 mm i.d., 1.8 μm particle size) column (Agilent, Santa Clara, CA, USA), protected with a Zorbax RRHD Eclipse Plus C18 (3.0 mm i.d., 1.8 µm particle size) guard column (Agilent, Santa Clara, CA, USA). The mobile phase was composed of a 10 mM ammonium formate buffer containing 0.05% of formic acid (solvent A) and MeOH (solvent B). Elution was carried out at a flow rate of 0.4 mL min−1 with chromatographic column thermostated at 35 °C. Elution started with 5% of solvent B, held 1 min. Solvent B was linearly increased to 30% in 3 min, then to 60% in 8 min and, finally, to 100% in 2 min, held for 2 min. Back to initial conditions was carried out in 2 min and held for 2 min for equilibration. The total run time was 20 min.
The LC system was coupled to a 6495 triple quadrupole mass spectrometer (MS/MS) equipped with an electrospray ionisation source operated in positive mode. The following settings were used: fragmentor, 166 V; capillary voltage, 4000 V; nebuliser pressure, 40 psi; sheath gas flow rate, 12 L min−1, sheath gas temperature, 250 °C; drying gas flow rate, 11 L min−1 and gas temperature, 350 °C.
Results and discussion
LC–MS/MS optimisation
LC–MS/MS parameters were optimised by injection of individual and mixture standard solutions of the selected compounds at 1 mg L−1. The type and composition of mobile phase solvents were optimised to achieve the highest compound ionisation to improve analytical signals and lower limits of detection. First, the aqueous phase (solvent A) was optimised using MeOH as organic solvent (solvent B). Ammonium formate and ammonium acetate, at different concentrations (from 2 to 10 mM) and with different additives (formic acid for ammonium formate and acetic acid for ammonium acetate (from 0 to 0.2% v/v)), were tested. The optimisation was carried out in both positive and negative modes. For all the compounds, the best results were obtained in positive mode. The precursor ions corresponded to the molecular ions after protonation. The highest intensities were provided by ammonium formate 10 mM containing 0.05% v/v of formic acid so that mixture was selected as aqueous mobile phase solvent. In all cases, the two most abundant product ions were monitored for each compound, one for quantification and the other for confirmation.
Then, the use of ACN as an organic solvent instead of MeOH was tested. As no improvement was observed, MeOH was selected due to its lower price and toxicity in comparison to ACN [33]. The flow rate was optimised in the range from 0.3 to 0.6 mL min−1 to reduce run time as much as possible at acceptable peak resolution. A flow rate of 0.4 mL min−1 was selected because it provided good resolution with low column pressure.
The optimised LC–MS/MS parameters for each compound are given in Table 1. The analyses were carried out using dynamic multiple reaction-monitoring mode (dMRM).
Method optimisation
The most significant parameters affecting UAE (type, acidification and volume of the extraction solvent, extraction time and number of extraction cycles) and d-SPE clean-up (type and amount of sorbent) were evaluated. Optimisation was carried out with faeces (0.5 g dw) spiked to a final concentration of 100 ng g−1 dw for each of the target compounds. The spiked faeces were incubated in the dark for 12 h to allow equilibration. Experiments were carried out in triplicate.
Extraction solvent optimisation
Four solvents (ACN, acetone, MeOH and hexane) were tested. Samples were ultrasonicated for 10 min using 3 mL of the tested solvent. Then, extracts were subjected to d-SPE clean-up by the addition of C18 (0.8 g). To calculate extraction recoveries, signals obtained from spiked faeces samples were compared with those from a matrix-matched standard at the same concentration.
As can be seen in Fig. 1, the best results were achieved with MeOH (mean recoveries were 75% except for tetracyclines), followed by acetone (49%), ACN (46%) and hexane (5%). The chemical structures of antibiotics are generally more complex and bigger than those of other pharmaceuticals. They have several ionisable functional groups and variable water solubilities and polarities (Table S1 in ESM). The low recoveries achieved for tetracyclines, with all the tested solvents, could be explained by the formation of complexes with bi- and trivalent cations present in the matrix, as was described previously [23, 34].
The second optimisation experiment comprised the study of the influence of the acidification of MeOH (up to 5% v/v of acetic acid). Acidified MeOH increased the signals from tetracyclines and their metabolite which can be explained by their acidic properties. However, the addition of acetic acid has a negative effect on neutral and basic antibiotics, mainly for AcSMX that suffer a recovery decrease from 30 to 35%. Sulfonamides, fluoroquinolones and antifolates (except 4-OH-TMP) presented the best extraction recoveries when pure MeOH was used whereas macrolides and their metabolites had not a common optimal extraction solvent. In addition, it has been reported that acid media, and the presence of buffers like citrate, can produce the epimerisation of tetracyclines [23], TC to EP-TC, resulting in alterations in the concentrations of each compound. Tetracyclines were poorly extracted. Nevertheless, no other additives were tested as pure MeOH provided good extraction recoveries for most of the antibiotics. It was selected as an extraction solvent.
Clean-up optimisation: type and amount of d-SPE sorbent
Clean-up was optimised in order to choose the most suitable sorbent, or sorbents, to remove interfering compounds without removing target compounds. Three sorbents (C18, PSA and Florisil®) and three sorbent amounts (0, 0.4 and 0.8 g) were simultaneously evaluated using a Box-Behnken experimental design (BBD). Samples were extracted by sonication for 10 min using 3 mL of MeOH. Extracts were subjected to the 15 clean-up experiments generated by the BBD matrix (see Table S2 in ESM). Clean-up efficiency was evaluated by comparing signals obtained after clean-up of spiked matrix extracts with those from a standard solution in MeOH:water (1:1, v/v) at the same concentration. Similar response surface plots were obtained from the five antibiotic families. In Fig. 2, it can be seen response surface plots corresponded to the average clean-up efficiency (%). Poor clean-up efficiency was obtained with PSA and Florisil®. The best results were obtained with 0.8 g of C18. Therefore, the conditions were selected for d-SPE extract clean-up.
UAE optimisation: solvent volume, extraction time and number of extraction cycles
Extraction time, solvent volume and number of extraction cycles were optimised by BBD. Three levels were evaluated for each variable: extraction time (5, 10 and 15 min), extraction solvent volume (3, 5 and 7 mL) and number of extraction cycles (1, 2 and 3). A total of 15 extraction experiments were performed (see Table S3 in ESM). After each extraction experiment, extract clean-up with 0.8 g of C18 was applied. Response surface plots, corresponding to mean method recovery (%), were constructed to better evaluate the effects of each variable and their interactions (Fig. 3). The most significant parameter was the number of extraction cycles. Three extraction cycles were necessary to quantitatively extract antibiotics and their metabolites. The best results for extraction time and MeOH volume were 10 min and 5 mL, respectively.
Response surface plots corresponding to mean method recovery when optimising the following pair of UAE factors: A number of extraction cycles vs extraction time; B number of extraction cycles vs MeOH volume; C extraction time vs MeOH volume. Faeces samples were spiked at 100 ng g−1 dw for each pollutant
Method validation
The method was validated in terms of matrix effect (ME), linearity, sensitivity, accuracy, precision and selectivity. ME was assessed by comparison of the calibration curve slopes in matrix-matched standards and in pure standard solutions in MeOH:water (1:1, v/v). Eight-point calibration curves were prepared in the range from method quantification limits (MQL) to 200 ng g−1 dw. Student’s t-test, at 95% of confidence, revealed significant differences between curve slopes which confirmed the presence of ME. Therefore, matrix-matched calibration was applied for the quantification of all the analytes. Their determination coefficients (R2) were higher than ≥ 0.994 for all the compounds (Table 2). ME was quantified by comparison of the peak area of the target compounds in matrix extract (Aextract), after subtracting the peak area obtained from non-spiked extracts (Ablank), and in pure solvent standard solutions (Astandard) following equation: ME (%) = (Aextract—Ablank—Astandard)/Astandard × 100). ME was quantified at three concentration levels: 5, 50, and 100 ng g−1 dw, except for 4-OH-TMP, TC and EP-TC which concentration levels were 75, 100 and 200 ng g−1 dw, respectively. Both matrix suppression and enhancement were obtained in faeces samples, especially for tetracycline family. RMX, CLM, DM-CLM, TMP, 4-OH-TMP, DM-TMP, SMX, SDZ, ERY and SMZ suffered matrix suppression while AZM, TC, EP-TC, AcSMX, SMX-GL, AcSDZ, NOR, ENR, CIP and AcSMZ suffered matrix enhancement effect in bird faeces matrix.
Sensitivity was assessed in terms of method detection limits (MDL) and MQL. These parameters were calculated as the concentrations provided signal to noise ratios of 3 and 10 for MDL and MQL, respectively. MQL values were lower than 2 ng g−1 dw except for TC (25 ng g−1 dw), 4-OH-TMP (50 ng g−1 dw) and EP-TC (50 ng g−1 dw). Table 2 shows obtained values. The extraction recoveries, accuracies, and precision of the method were evaluated using spiked faeces samples at three concentration levels in triplicate. Spike concentrations were 5 ng g−1 dw (low), 50 ng g−1 dw (medium) and 100 ng g−1 dw (high), for compounds with MQL values higher than 2 ng g−1 dw; and 75 ng g−1 dw (low), 100 ng g−1 dw (medium) and 200 ng g−1 dw (high) for TC, EP-TC and 4-OH-TMP. Extraction recoveries (R) were calculated by comparison of the peak areas obtained from the spiked samples (Asample) with those from spiked extracts (Aextract), after blank correction (Ablank) applying equation: R (%) = (Asample—Ablank)/(Aextract—Ablank) × 100. Accuracy (A), expressed as relative recovery, was determined by comparison of the concentration obtained from spiked samples using matrix-matched calibration curves (Cspiked sample), after blank correction (Cblank), with the spike concentration (Cspike concentration) applying equation: A (%) = (Cspiked sample—Cblank) × 100/R (%) × 100/Cspike concentration. Results are shown in Table 2. Accuracies at the three spike concentrations were in the range from 42.4 to 104.2% for macrolides, 50.9–118.8% for fluoroquinolones, 60.1–126.5% for tetracyclines, 56.9–101.7% for antifolates and 56.0–106.3% for sulfonamides. Precision was calculated as inter-day repeatability and expressed as relative standard deviation (RSD, %). RSD values were below 24% for all compounds at the three spike concentrations. Method selectivity was evaluated by comparison of the chromatograms of procedural blanks and spiked faeces samples. No interference was observed at the retention times of the target compounds. In Fig. S1 (ESM), it can be seen a LC–MS/MS chromatogram of a faeces sample spiked at 75 ng g−1 dw.
Method comparison
In Table 3, it is summarised a comparison between the operational and analytical parameters of the proposed method and those reported in the literature for the determination of the target compounds in excreta. As mentioned in “Introduction,” such methods were developed for the determination of veterinary antibiotics in excreta and manure from farm animals, mainly mammals, not for environmentally relevant antibiotics in wildlife birds. Wild animals, and particularly synanthropic birds, are overlooked but key agents in the epidemiology of clinically important antibiotic-resistant bacteria [35, 36], so a specific analytical method for the determination of environmentally relevant antibiotics in wild bird faeces is highly required. The proposed extraction methods are mainly based on UAE [25, 28,29,30, 37] and on solid–liquid extraction (SLE) [22,23,24, 26, 38] followed by extract clean-up by SPE [21, 22, 26, 27, 29, 37]. QuEChERS (quick, easy, cheap, effective, rugged and safe) method [39] and pressurised liquid extraction (PLE) [21] have been also proposed. The proposed method allows several operational advantages such as a low sample mass (0.5 g), which is mandatory for the analysis of bird faeces; low-cost instrumentation in comparison to the PLE method; lower solvent consumption (15 mL) in comparison to QuECheRS, SPE clean-up and some of the proposed UAE and SLE methods (up to 144 and 317 mL, respectively); and no plastic waste generation in comparison to clean-up by SPE cartridges or QuECheRS method. Accuracy values are similar or even better than the above-mentioned methods. MDLs values achieved are lower (from 0.01 to 1.5 ng g−1 dw for 90% of the target compounds) than those reported by other authors (Table 3) especially in comparison to methods developed for antibiotics from different families for which MDLs up to 500 ng g−1 dw have been reported [39]. For instance, the method proposed by Pokrant et al. [40], for the determination of veterinary antibiotics in broiler chicken faeces, achieved MDLs in the range from 17.5 to 37. 4 ng g−1 in spite of a higher sample amount being treated (1 g) in comparison to the proposed method (0.5 g). Such MDLs were suitable for the determination of antibiotics after administration to chickens as their concentrations in faeces were in the range of 68 to 2058 ng g−1. Nevertheless, lower MDLs are required for the determination of environmentally relevant antibiotics in wild bird faeces. Furthermore, the proposed method allows the determination of 8 metabolites whereas only four of the published methods include the determination of metabolites of antibiotics but just two or three metabolites and from one or two families of antibiotics (sulfonamides [23]; tetracyclines [23, 27, 40]; or fluoroquinolones [37]). These facts are of special relevance as concentrations of pharmaceuticals in wild bird faeces are expected to be lower than in livestock and poultry excrements as antibiotics are intendedly administered to farm animals; and because bird animals can be directly exposed not only to parent compounds but also to metabolites present in the environment.
Method application
The developed method was applied to the determination of the target compounds in 27 faeces samples from three waterbirds species wintering in Spain: Ciconia ciconia (n = 15), Larus fuscus (n = 8) and Chroicocephalus ridibundus (n = 4). The results obtained are shown in Table S4 (ESM). A summary of the obtained results can be seen in Table 4. Nine parent compounds and three metabolites were found in analysed samples. Parent compounds were more frequently detected and at similar or higher concentrations than their metabolites, except for TC. The metabolite of TC (EP-TC)) was detected in just four faeces’ samples, all of them from the white stork Ciconia ciconia whereas TC was detected in no sample. It has been described that tetracyclines are instable compounds that can suffer abiotic degradation in the environment, conditioned by temperature, pH, light and redox conditions, being EP-TC one of the main degradation products [41]. Therefore, the detection of EP-TC in four faeces samples, whereas TC was not detected, could be due to a high TC metabolisation in white stork Ciconia ciconia or to a higher ingestion of EP-TC released to the environment through wastewater or generated as a degradation product in the environment and landfills. Nevertheless, more samples should be analysed to obtain concluding results. RXM, NOR and TMP were detected in all the analysed samples and bird species. AZM was detected in all samples, except Chroicocephalus ridibundus samples. Fluoroquinolones and macrolides were the families most frequently detected. The highest concentrations belonged to NOR (up to 199.27 ng g−1 in Ciconia ciconia faeces sample 9), SMX (up to 300.62 ng g−1 dw in Ciconia ciconia faeces sample 3) and AcSMX (up to 148.02 ng g−1 dw in Larus fuscus faeces sample 8). ERY, TC, 4-OH-TMP, SMX-GL, SMZ, AcSMZ and AcSDZ were not detected in any of the analysed samples.
Concentration levels measured are consistent with data available in faeces and manure samples from farm animals. For instance, Wang et al. [21] reported concentrations of RXM and SDZ up to 5.1 and 23.7 ng g−1 dw, respectively, in livestock and poultry excrements. Berendsen et al. [22] reported concentrations of CIP up to 13 ng g−1 dw in cattle faeces and Argüeso-Mata et al. [25] found concentrations of SMX up to 70 ng g−1 dw in pig manure. The high concentrations of antibiotics in the studied birds can be explained by their omnivorous and opportunistic habits. White storks (Ciconia ciconia), lesser black-backed gulls (Larus fuscus) and black-headed gulls (Chroicocephalus ridibundus) commonly use landfills and wastewaters for feeding, which have been reported to constitute an important reservoir of antibiotics [42]. This fact is consistent with results from a previous work in which it has been observed that faeces from storks and gulls contained a significantly higher abundance of antibiotic resistance genes than faeces from geese and cranes that feed in more natural habitats [9]. Moreover, the studied birds can acquire antibiotics by ingestion of preys where antibiotics can be bioaccumulated as reported for many wild-living aquatic organisms [43]. In addition, a concentration effect can occur in faeces, with respect to the amounts of the antibiotics in water and food ingested by the migratory birds.
Conclusions
An analytical method for the determination of five antibiotic families and their main metabolites in bird faeces samples has been optimised and validated. To the date and to the best of our knowledge, the proposed method constitutes the first one for multiresidue determination of different families of antibiotics and their main metabolites in wild bird faeces samples. This fact is of especial relevance not only because the composition of wild bird faeces is different from poultry excrements but also because antibiotics to which wild birds are exposed can be different to antibiotics administered to poultry.
In addition, this is the first method combining UAE and d-SPE for the determination of antibiotics in faeces samples. The method allowed good linearity (R2 ≥ 0.994), accuracy close to 100%, adequate precision (RSD < 24%) and low MQLs (< 2 ng g−1 dw) for most of the compounds. Recoveries and MDLs were similar or improved than those reported for other excrement matrices but requires lower solvent volumes and sample amounts.
The analysis of 27 faeces samples from three common migratory waterbirds species revealed the presence of 9 out of the 12 antibiotics and 3 of their main metabolites. The proposed method can be a useful tool not only to monitor environmental risks for wild waterbirds but also (i) to assess biovectoring and quantification of antibiotic load into specific environments; (ii) to evaluate the environmental risk caused by the environmental dissemination of residues of antibiotics; and (iii) as a “tool” to reveal the overuse of antibiotics and their environmental load. This information is increasingly important due to the need for a better integration of wildlife into the current One Health approach for antibiotic resistance surveillance and control.
References
Knapp CW, Zhang W, Sturm BSM, Graham DW. Differential fate of erythromycin and beta-lactam resistance genes from swine lagoon waste under different aquatic conditions. Environ Pollut. 2010;158(5):1506–12. https://doi.org/10.1016/j.envpol.2009.12.020.
Milić N, Milanović M, Letić NG, Sekulić MT, Radonić J, Mihajlović I, et al. Occurrence of antibiotics as emerging contaminant substances in aquatic environment. Int J Environ Health Res. 2013;23(4):296–310. https://doi.org/10.1080/09603123.2012.733934.
Bilal M, Mehmood S, Rasheed T, Iqbal HMN. Antibiotics traces in the aquatic environment: persistence and adverse environmental impact. Curr Opin Environ Sci Heal. 2020;13:68–74. https://doi.org/10.1016/j.coesh.2019.11.005.
Ghirardini A, Grillini V, Verlicchi P. A review of the occurrence of selected micropollutants and microorganisms in different raw and treated manure – environmental risk due to antibiotics after application to soil. Sci Total Environ. 2020;707:136118. https://doi.org/10.1016/j.scitotenv.2019.136118.
Zhuang M, Achmon Y, Cao Y, Liang X, Chen L, Wang H, et al. Distribution of antibiotic resistance genes in the environment. Environ Pollut. 2021;285:117402. https://doi.org/10.1016/j.envpol.2021.117402.
Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, et al. Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol. 2015;13(5):310–7. https://doi.org/10.1038/nrmicro3439.
Cristóbal-Azkarate J, Dunn JC, Day JMW, Amábile-Cuevas CF. Resistance to antibiotics of clinical relevance in the fecal microbiota of Mexican wildlife. PLoS ONE. 2014;9(9):3–10. https://doi.org/10.1371/journal.pone.0107719.
Hernando-Amado S, Coque TM, Baquero F, Martínez JL. Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nat Microbiol. 2019;4(9):1432–42. https://doi.org/10.1038/s41564-019-0503-9.
Jarma D, Sánchez MI, Green AJ, Peralta-Sánchez JM, Hortas F, Sánchez-Melsió A, et al. Faecal microbiota and antibiotic resistance genes in migratory waterbirds with contrasting habitat use. Sci Total Environ. 2021;783:146872. https://doi.org/10.1016/j.scitotenv.2021.146872.
Martín-Vélez V, Hortas F, Taggart MA, Green AJ, ÓHanlon NJ, Sánchez MI. Spatial variation and biovectoring of metals in gull faeces. Ecol Indic. 2021;125:107534. https://doi.org/10.1016/j.ecolind.2021.107534
Murray CG, Hamilton AJ. Perspectives on wastewater treatment wetlands and waterbird conservation. J Appl Ecol. 2010;47(5):976–85. https://doi.org/10.1111/j.1365-2664.2010.01853.x.
Rabbia V, Bello-Toledo H, Jiménez S, Quezada M, Domínguez M, Vergara L, et al. Antibiotic resistance in Escherichia coli strains isolated from Antarctic bird feces, water from inside a wastewater treatment plant, and seawater samples collected in the Antarctic Treaty area. Polar Sci. 2016;10(2):123–31. https://doi.org/10.1016/j.polar.2016.04.002.
Pokrant E, Maddaleno A, Lobos R, Trincado L, Lapierre L, San Martín B, et al. Assessing the depletion of lincomycin in feathers from treated broiler chickens: a comparison with the concentration of its residues in edible tissues. Food Addit Contam - Part A Chem Anal Control Expo Risk Assess. 2019;36(11):1647–53. https://doi.org/10.1080/19440049.2019.1662952.
Gajda A, Nowacka-Kozak E, Gbylik-Sikorska M, Posyniak A. Feather analysis as a non-invasive alternative to tissue sampling for surveillance of doxycycline use on poultry farms. Poult Sci. 2019;98(11):5971–80. https://doi.org/10.3382/ps/pez394.
Gajda A, Nowacka-Kozak E, Gbylik-Sikorska M, Posyniak A. Multi-residues UHPLC–MS/MS analysis of 53 antibacterial compounds in poultry feathers as an analytical tool in food safety assurance. J Chromatogr B Anal Technol Biomed Life Sci. 2019;1104:182–9. https://doi.org/10.1016/j.jchromb.2018.11.020.
Maddaleno A, Pokrant E, Yanten F, San Martin B, Cornejo J. Implementation and validation of an analytical method for lincomycin determination in feathers and edible tissues of broiler chickens by liquid chromatography tandem mass spectrometry. J Anal Methods Chem. 2019;2019:4569707. https://doi.org/10.1155/2019/4569707.
Chiesa LM, Nobile M, Panseri S, Arioli F. Suitability of feathers as control matrix for antimicrobial treatments detection compared to muscle and liver of broilers. Food Control. 2018;91:268–75. https://doi.org/10.1016/j.foodcont.2018.04.002.
Cornejo J, Pokrant E, Carvallo C, Maddaleno A, San MB. Depletion of tylosin residues in feathers, muscle and liver from broiler chickens after completion of antimicrobial therapy. Food Addit Contam - Part A Chem Anal Control Expo Risk Assess. 2018;35(3):448–57. https://doi.org/10.1080/19440049.2017.1401740.
Jansen LJM, Bolck YJC, Rademaker J, Zuidema T, Berendsen BJA. The analysis of tetracyclines, quinolones, macrolides, lincosamides, pleuromutilins, and sulfonamides in chicken feathers using UHPLC-MS/MS in order to monitor antibiotic use in the poultry sector. Anal Bioanal Chem. 2017;409(21):4927–41. https://doi.org/10.1007/s00216-017-0445-0.
Berendsen BJA, Bor G, Gerritsen HW, Jansen LJM, Zuidema T. The disposition of oxytetracycline to feathers after poultry treatment. Food Addit Contam - Part A Chem Anal Control Expo Risk Assess. 2013;30(12):2102–7. https://doi.org/10.1080/19440049.2013.843026.
Wang J, Xu J, Ji X, Wu H, Yang H, Zhang H, et al. Determination of veterinary drug/pesticide residues in livestock and poultry excrement using selective accelerated solvent extraction and magnetic material purification combined with ultra-high-performance liquid chromatography–tandem mass spectrometry. J Chromatogr A. 2020;1617:460808. https://doi.org/10.1016/j.chroma.2019.460808.
Berendsen BJA, Wegh RS, Memelink J, Zuidema T, Stolker LAM. The analysis of animal faeces as a tool to monitor antibiotic usage. Talanta. 2015;132:258–68. https://doi.org/10.1016/j.talanta.2014.09.022.
Spielmeyer A, Ahlborn J, Hamscher G. Simultaneous determination of 14 sulfonamides and tetracyclines in biogas plants by liquid-liquid-extraction and liquid chromatography tandem mass spectrometry. Anal Bioanal Chem. 2014;406(11):2513–24. https://doi.org/10.1007/s00216-014-7649-3.
Qiu J, Zhao T, Liu Q, He J, He D, Wu G, et al. Residual veterinary antibiotics in pig excreta after oral administration of sulfonamides. Environ Geochem Health. 2016;38(2):549–56. https://doi.org/10.1007/s10653-015-9740-x.
Argüeso-Mata M, Bolado S, Jiménez JJ, López-Serna R. Determination of antibiotics and other veterinary drugs in the solid phase of pig manure. Chemosphere. 2021;275:130039. https://doi.org/10.1016/j.chemosphere.2021.130039.
Widyasari-Mehta A, Suwito HRKA, Kreuzig R. Laboratory testing on the removal of the veterinary antibiotic doxycycline during long-term liquid pig manure and digestate storage. Chemosphere. 2016;149:154–60. https://doi.org/10.1016/j.chemosphere.2016.01.094.
Shelver WL, Varel VH. Development of a UHPLC-MS/MS method for the measurement of chlortetracycline degradation in swine manure. Anal Bioanal Chem. 2012;402(5):1931–9. https://doi.org/10.1007/s00216-011-5637-4.
Xu M, Li H, Li S, Li C, Li J, Ma Y. The presence of tetracyclines and sulfonamides in swine feeds and feces: dependence on the antibiotic type and swine growth stages. Environ Sci Pollut Res. 2020;27(34):43093–102. https://doi.org/10.1007/s11356-020-10266-5.
Ho YB, Zakaria MP, Latif PA, Saari N. Simultaneous determination of veterinary antibiotics and hormone in broiler manure, soil and manure compost by liquid chromatography-tandem mass spectrometry. J Chromatogr A. 2012;1262:160–8. https://doi.org/10.1016/j.chroma.2012.09.024.
Gorissen B, Reyns T, Devreese M, De Backer P, Van Loco J, Croubels S. Determination of selected veterinary antimicrobials in poultry excreta by UHPLC-MS/MS, for application in Salmonella control programs. Anal Bioanal Chem. 2015;407(15):4447–57. https://doi.org/10.1007/s00216-014-8449-5.
Singer MA. Do mammals, birds, reptiles and fish have similar nitrogen conserving systems? Comp Biochem Physiol - B Biochem Mol Biol. 2003;134(4):543–58. https://doi.org/10.1016/S1096-4959(03)00027-7.
Van der Hoeven-Hangoor E, Van de Linde IB, Paton ND, Verstegen MWA, Hendriks WH. Effect of different magnesium sources on digesta and excreta moisture content and production performance in broiler chickens. Poult Sci. 2013;92(2):382–91. https://doi.org/10.3382/ps.2012-02404.
Joshi DR, Adhikari N. An overview on common organic solvents and their toxicity. J Pharm Res Int. 2019;28(3):1–18. https://doi.org/10.9734/jpri/2019/v28i330203.
Mejías C, Martín J, Santos JL, Aparicio I, Alonso E. Occurrence of pharmaceuticals and their metabolites in sewage sludge and soil: a review on their distribution and environmental risk assessment. Trends Environ Anal Chem. 2021;30:e00125. https://doi.org/10.1016/j.teac.2021.e00125.
Dolejska M, Literak I. Wildlife is overlooked in the epidemiology of medically important antibiotic-resistant bacteria. Antimicrob Agents Chemother. 2019;63(8):e01167-e1219. https://doi.org/10.1128/AAC.01167-19.
Jarma D, Sánchez MI, Green AJ, Peralta-Sánchez JM, Hortas F, Sánchez-Melsió A, Borrego CM. Faecal microbiota and antibiotic resistance genes in migratory waterbirds with contrasting habitat use. Sci Total Environ. 2021;783:146872. https://doi.org/10.1016/j.scitotenv.2021.146872.
Slana M, Žigon D, Sollner-Dolenc M. Enrofloxacin degradation in broiler chicken manure under field conditions and its residuals effects to the environment. Environ Sci Pollut Res. 2017;24(15):13722–31. https://doi.org/10.1007/s11356-017-8722-1.
Bailey C, Spielmeyer A, Hamscher G, Schüttrumpf H, Frings RM. The veterinary antibiotic journey: comparing the behaviour of sulfadiazine, sulfamethazine, sulfamethoxazole and tetracycline in cow excrement and two soils. J Soils Sediments. 2016;16(6):1690–704. https://doi.org/10.1007/s11368-016-1370-0.
Wang K, Wang X, Xu Z, Yang S. Simultaneous determination of multi-class antibiotics and steroid hormones drugs in livestock and poultry faeces using liquid chromatography–quadrupole time-of-flight mass spectrometry. Food Addit Contam - Part A Chem Anal Control Expo Risk Assess. 2020;37(9):1467–80. https://doi.org/10.1080/19440049.2020.1776900.
Pokrant E, Trincado L, Yévenes K, Terraza G, Maddaleno A, San Martín B, Zabala S, Hidalgo H, Lapierre L, Cornejo J. Determination of five antimicrobial families in droppings of therapeutically treated broiler chicken by high-performance liquid chromatography-tandem mass spectrometry. Poult Sci. 2021;100:101313. https://doi.org/10.1016/j.psj.2021.101313.
Wu X, Wei Y, Zheng J, Zhao X, Zhong W. The behavior of tetracyclines and their degradation products during swine manure composting. Bioresour Technol. 2011;102:5924–31. https://doi.org/10.1016/j.biortech.2011.03.007.
Zhang R, Yang S, An Y, Wang Y, Lei Y, Song L. Antibiotics and antibiotic resistance genes in lanfills: a review. Sci Total Environ. 2022;806:150647. https://doi.org/10.1016/j.scitotenv.2021.150647.
Świacka K, Maculewicz J, Kowalska D, Caban M, Smolarz K, Świeżak J. Presence of pharmaceuticals and their metabolites in wild-living aquatic organisms – current state of knowledge. J Hazard Mater. 2022;424:127350. https://doi.org/10.1016/j.jhazmat.2021.127350.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. This work was financially supported by the Ministerio de Ciencia e Innovación from Spanish Government (Project No. PID2020-117641RB-I00 and No. PID2019-108962 GB-C21).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mejías, C., Martín, J., Santos, J.L. et al. Development and validation of a highly effective analytical method for the evaluation of the exposure of migratory birds to antibiotics and their metabolites by faeces analysis. Anal Bioanal Chem 414, 3373–3386 (2022). https://doi.org/10.1007/s00216-022-03953-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-022-03953-4