Abstract
Paper has been widely employed as cheap material for the development of a great number of sensors such as pregnancy tests, strips to measure blood sugar, and COVID-19 rapid tests. The need for new low-cost analytical devices is growing, and consequently the use of these platforms will be extended to different assays, both for the final consumer and within laboratories. This work describes a paper-based electrochemical sensing platform that uses a paper disc conveniently modified with recognition molecules and a screen-printed carbon electrode (SPCE) to achieve the detection of gluten in a deep eutectic solvent (DES). This is the first method coupling a paper biosensor based on aptamers and antibodies with the DES ethaline. Ethaline proved to be an excellent extraction medium allowing the determination of very low gluten concentrations. The biosensor is appropriate for the determination of gluten with a limit of detection (LOD) of 0.2 mg L−1 of sample; it can detect gluten extracted in DES with a dynamic range between 0.2 and 20 mg L−1 and an intra-assay coefficient of 10.69%. This approach can be of great interest for highly gluten-sensitive people, who suffer from ingestion of gluten quantities well below the legal limit, which is 20 parts per million in foods labeled gluten-free and for which highly sensitive devices are essential.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Paper displays interesting physical and physicochemical properties, such as adsorption properties, capillary action, and high surface-to-volume ratio, and allows immobilization of biomolecules [1]. It has been applied in many different research fields, such as in the development of sensors, microfluidic devices, and point-of-care(POC) diagnostic tools [2]. In recent decades, POC tests based on paper have been developed for glucose and other important bioactive molecules [3, 4]. Currently, paper continues to be employed as material for the production of widely used sensors such as pregnancy tests, strips to measure blood sugar, and COVID-19 rapid tests [5, 6].
Besides paper strips, patterned paper has also been used as a platform for the implementation of portable, low-cost bioassays aimed at use in developing countries [7, 8]. In addition, electrochemical detection for paper-based microfluidics was also proposed for the determination of low levels of analytes in biological samples and complex sample matrixes [9].
The need for new low-cost analytical devices is growing, and the use of these platforms will be extended to different assays both for the final consumer and within laboratories [10, 11]. Among the most relevant points in the use of this material, there are advantages such as biocompatibility and biodegradability, low cost, and ease of production [12]. These aspects have led to a growing interest in the development of paper-based analytical devices (PADs), such as smart labels [13], gas sensors [14, 15], and sensors combining electrochemical and visual readouts [16]. PADs have successfully found application in diagnostics [4], environmental monitoring [17], and food control [18].
To date, paper-based gluten sensors such as lateral flow devices are commercially available, indicating the presence or absence of gluten, with a limit of detection (LOD) of around 4 mg L−1. They can be used for potentially contaminated surfaces and to check for gluten contamination of raw or processed materials [19], but they are not suitable for sensitive gluten quantification.
As is well known, celiac disease is triggered by the ingestion of gluten in people predisposed to the disease [20]. In the future, it will be increasingly necessary for consumers to monitor food directly at home. Thus, the development of low-cost platforms that are easy to use and highly sensitive is of growing interest [18].
Gluten is composed of a complex mixture of water-insoluble storage proteins; among them, gliadin is commonly used as the analytical target to quantify gluten in food. The most commonly used solvent in gluten quantification methods is a 60% (v/v) ethanol-water solution; however, this method is not able to completely extract gluten from processed food [21]. Reducing and disaggregating agents have also been used in combination with alcohol solutions to overcome this problem [22, 23]. Nevertheless, both 2-mercaptoethanol and denaturants used in the extraction cocktails can interfere in the subsequent protein recognition, affecting the quantification results [24]. Thus, substantial sample dilutions are usually needed. The problem regarding the complete extraction of gluten proteins from food makes the determination of gluten a continuing challenge and an open topic in which research advances are needed [25].
Recently, an alternative method of extraction using a deep eutectic solvent (DES) was proposed [26]. This approach allows the direct measurement of the extracted sample in the DES ethaline (choline chloride:ethylene glycol, 1:2), exploiting the biocompatibility of the eutectic solvent with molecules such as DNA and antibodies.
DESs are formed thanks to the interaction between a hydrogen bond donor (HBD) and a hydrogen bond acceptor (HBA) [27]. They present low vapor pressure and a high ability to dissolve molecules of different nature; they are green, easy to produce, and low-cost [28, 29]. For these reasons, the use of DESs is expanding in different fields [30,31,32,33], in fact, in recent years, they have been applied in the extraction of various molecules [34,35,36] and in different research areas including organic synthesis, electrochemistry, and biocatalysis [37,38,39].
Here we describe a paper-based electrochemical sensing platform that uses a paper disc conveniently modified with recognition molecules and a screen-printed electrode to achieve the detection of gluten in DES. This analytical platform allows the use of a single screen-printed carbon electrode (SPCE) for the analysis of several paper discs. This provides the possibility for reduced use of SPCEs and a considerable decrease in the cost of each measurement.
This sandwich-type electrochemical biosensor exploits an aptamer and antibody pair for the detection of gluten. Aptamer-antibody sandwich assays show enhanced sensitivity and specificity [40, 41]. For this reason, a Gli4-T aptamer was employed as the capturing element, while a 401/21 antibody was used as detection probe. This platform was designed to provide a low-cost and sensitive tool for the fraction of highly gluten-sensitive people, who suffer from ingestion of gluten quantities well below the legal limit which is 20 parts per million in foods labeled gluten-free and for which highly sensitive devices are essential [42].
Materials and methods
Chemicals and reagents
All chemicals used were of analytical reagent grade and were employed without further purification. HPLC-purified 5′-tagged (-NH2) aptamers were obtained from Tema Ricerca (Bologna, Italy). The Gli4 complete aptamer sequence and the truncated sequence are shown in Table 1. The 401/21 antibody and enzyme-linked immunosorbent assay (ELISA) kit were supplied by Bio-Check (UK). Gliadin standard was kindly provided by the Prolamin Working Group (PWG). Ethaline was supplied by Scionix Ltd. (London, UK) and employed as received (Table 2). Salts for buffer solutions, bovine serum albumin (BSA), 1 M Tris/HCl pH 7.4, phosphate-buffered saline (PBS) 10×, sodium alginate, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC), N-hydroxysuccinimide (NHS), and a 3,3′,5,5′-tetramethylbenzidine (TMB) liquid substrate system for ELISA solution were obtained from Sigma-Aldrich (Italy). With regard to filter paper, 50 cm × 50 cm foils, 67 g/m2, 0.13 mm thick, were purchased from Labor (Cordenons, Italy) and 0.18 mm-thick paper from Whatman (Maidstone, UK).
High-purity deionized water (purified by an Elgastat UHQ PS system, Elga, High Wycombe, UK) was used for all washing operations.
Paper-based biosensor assembly
Filter paper was cut into discs of 6 mm. As shown in Fig. 1, discs were soaked in 10 μL of sodium alginate (0.05% w/v) at 4 °C overnight. After a washing step, the polysaccharide-modified paper was ready for use. Then, 10 μL of a mixed solution (EDC 0.4 M and NHS 0.2 M) was used to activate the carboxylic groups of the polysaccharide. The 5′ amined aptamer (1 μM) was covalently immobilized on the paper during 2 h incubation, and then the surface was blocked for 15 min with a solution of 1% BSA. Subsequently, solutions of increasing concentration (between 0.1 and 10 mg L−1) of PWG gliadin in buffer (Tris 50 mM, NaCl 250 mM, MgCl2 5 mM) or alternatively in ethaline were incubated for 20 min. Next, a solution of the anti-gluten antibody was incubated for 10 min. After each step, discs were washed with buffer solution. Finally, electrochemical transduction was performed by adding 40 μL of TMB solution, and after 3 min of the enzymatic reaction the paper disc was transferred onto the SPCE and measured by chronoamperometry at 0 V (Figure1).
Electrochemical measurements
All electrochemical measurements were performed with a µAutolabIII/FRA2 potentiostat controlled by Nova 2.1 software (EcoChemie, the Netherlands). Disposable screen-printed carbon electrodes (SPCE) ref. C110 were purchased from Metrohm DropSens (Metrohm Italiana s.r.l., Varese, Italia).
Sample preparation and extraction procedure
To prevent contamination, samples were prepared in a laboratory separated from where analyses were performed. Gluten was extracted with ethaline from corn flakes, and a gluten-free flour purchased from a local supermarket.
A 0.1 g sample of the obtained powder was extracted in vials with 1 mL of pure DES [43]. Vials were shaken in a vortex for 2 min, and then they were left at 55 °C for 45 min. Next, samples were shaken again for 2 min and centrifuged for 10 min at 5000 rpm. Depending on the level of gluten, the supernatant was applied directly on the electrode surface or properly diluted in 100% DES before the analysis.
Results and discussion
Modification of SPCEs with paper
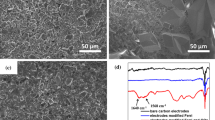
Initially, the modification of the SPCE with the paper disc was evaluated with cyclic voltammetry at different scan rates (25, 50, 100 mV/s) using the redox probe ferrocyanide 2 mM in PBS buffer and KCl 3 mM as supporting electrolyte. Cyclic voltammograms recorded at the bare SPCE and filter paper-modified SPCE and Whatman paper-modified SPCE are presented in Fig. 2. A well-defined and quasi-reversible redox process was found for the redox couple [Fe(CN)6]4−/ [Fe(CN)6]3− for all the setups, with a peak-to-peak separation (ΔEp) recorded at 50 mV/s of 152 mV on bare SPCE, 91 mV on filter paper, and 110 mV on Whatman paper. These results show that the electron transfer on paper-modified SPCE is very similar to that recorded at bare SPCE, suggesting that good electrolytic contact was achieved. However, the measurements performed on Whatman paper showed a background noise at high scanning speeds.
Gliadin quantification by paper-based biosensor
The principle of the biosensor is illustrated in Fig. 1. In this approach, the truncated aptamer Gli4-T was employed as capture element while the 401/21 antibody was employed as the signaling element. In a previous work, the process of truncation of non-essential nucleotides was reported to improve the accessibility of the target to the aptamer, allowing the formation of a stronger aptamer-target complex [44]. Gli4-T is the truncated aptamer derived from Gli4 which preserves a motif capable of effectively binding gliadin (Fig. 3) [45, 46]. Gli4-T has a dissociation constant (Kd) of 148 ± 8 nM in aqueous buffer, and a Kd of 515 ± 144 nM in ethaline [43].
The paper-based sensor was designed using common laboratory filter paper cut into discs of 6 mm. A polysaccharide with multiple functional groups was employed as surface modifier for paper on which the aptamer was covalently immobilized [47]. Sodium alginate was dropped onto the paper, and the carboxyl groups were then activated by EDC/NHS and subsequently the amino-modified aptamer was immobilized. After blocking the surface with BSA, PWG-gliadin solutions were tested in the range of 0.1–10 mg L−1. Lastly, the antibody labeled with horseradish peroxidase (HRP) enzyme was incubated and TMB substrate added. The enzyme catalyzed the oxidation of TMB in the presence of hydrogen peroxide, then the chronoamperometric detection was performed at 0 V. Both filter paper and Whatman paper were tested in the development of the assay. As can be observed in Fig. 4, at lower concentrations of analyte there was little difference in the performance, but at higher concentrations of gliadin (1 mg L−1), the common filter paper showed a higher signal ratio, proving to be the best option for the assay development.
For the sake of comparison, calibration curves were conducted both in aqueous buffer and in ethaline. As can be seen in Fig. 5, both calibration curves fitted to the Hill function, with a correlation of 0.999 in aqueous buffer and 0.997 in ethaline.
The calibration in DES allowed to expand the measurement to 10 mg L−1, while the calibration in aqueous buffer already reached the saturation at a concentration of 2 mg L−1 of gliadin. The response of the biosensor was determined in a range from 0.1 to 1 mg L−1 of gliadin in aqueous buffer and from 0.1 to 10 mg L−1 in ethaline.
The reproducibility of the method was very good; the calculated intra-assay CV % in aqueous buffer was 6.64 considering all tested concentrations on three different days, while in ethaline it was 10.69%.
The results obtained with this sandwich sensor proved to be interesting for the sensitive, low-cost detection of gluten. In addition, the obtained LOD, calculated as three times the standard deviation of the blank signal divided by the slope calculated for the linear dynamic range, is 0.2 mg L−1 of gluten, which successfully competes with the assays described in the literature and on the market, as can be seen in Table 3.
Application of the assay to food samples extracted in DES
The paper-based biosensor here developed can be applied in the determination of gluten in gluten-free foods to provide more information and safety to celiac people. To demonstrate its applicability to gluten quantification, two real food samples were analyzed. We tested a gluten-free flour sample and a corn flakes sample using ethaline for the extraction. Accordingly, we also performed the quantification with an ELISA, and the results are compared in Table 4.
The quantification of gluten in the flour sample with the paper-based method gave a gluten content of 2.68 ± 0.01 mg L−1, a result comparable to the one obtained with the classic ELISA (2.3 ± 0.1 mg L−1) of gluten. Moreover, the sensor also worked well for the corn flakes sample with a higher gluten content (21.33 ± 0.08 mg L−1) in good agreement with the results obtained from the ELISA test (20.50 ± 0.09 mg L−1). These results proved that this method can be successfully used for the reliable quantification of gluten in foods labeled as gluten-free and can be a valid alternative to current methods. Moreover, to ensure the effectiveness and applicability of the method, we evaluated the possible interference caused by other proteins. For this purpose, chickpea flour with high protein content was used. After extraction with ethaline, the sample was tested with the paper-based sensor. This naturally gluten-free sample gave comparable signals as blanks, as can be seen in Table 5, demonstrating that this method shows no interference from the presence of other proteins in the sample.
Conclusions
In this work we have designed a simple, low-cost, and easy-to-use biosensor. This is the first method coupling a paper biosensor based on aptamers and antibodies with a DES. We coupled a paper disc with recognition biomolecules for the electrochemical determination of gluten extracted in DES, providing a cheap detection device in comparison with traditional methods. This approach could enable a substantial advance in the analysis of gluten, being very economical and sensitive at the same time. Very important aspects to take into consideration are the use of biodegradable materials such as paper and DES and the advantage of using a single SPCE for a whole set of measures. Moreover, this biosensor has proven to be appropriate for the determination of very low concentrations of gluten, with a LOD of 0.2 mg L−1 of sample. The ELISA R5 Mendez method remains the only method recommended by the Codex for gluten analysis; however, all advances in this area can bring important improvements in the quantification of gluten. This approach can be of great interest for highly gluten-sensitive people who suffer from ingestion of gluten quantities well below the legal limit and for whom highly sensitive devices are essential.
Data availability
Not applicable.
Code availability
Not applicable.
References
Liu S, Su W, Ding X. A Review on Microfluidic Paper-Based Analytical Devices for Glucose Detection. Sensors. 2016;16:2086.
Costa-Rama E, Fernández-Abedul MT. Paper-Based Screen-Printed Electrodes: A New Generation of Low-Cost Electroanalytical Platforms. Biosensors. 2021;11:51.
Turner APF. Biosensors: sense and sensibility. Chem Soc Rev. 2013;42:3184.
Hu J, Wang S, Wang L, Li F, Pingguan-Murphy B, Lu TJ, et al. Advances in paper-basedpoint-of-care diagnostics. Biosens Bioelectron. 2014;54:585–97.
Arduini F, Cinti S, Scognamiglio V, Moscone D. Chapter Eleven - Paper-Based Electrochemical Devices in Biomedical Field: Recent Advances and Perspectives. In: Palchetti I, Hansen P-D, Barceló D, editors. Comprehensive Analytical Chemistry: Elsevier; 2017. p. 385–413.
Peng T, Sui Z, Huang Z, Xie J, Wen K, Zhang Y, et al. Point-of-care test system for detection of immunoglobulin-G and -M against nucleocapsid protein and spike glycoprotein of SARS-CoV-2. Sens Actuators B Chem. 2021;331:129415.
Martinez AW, Phillips ST, Butte MJ, Whitesides GM. Patterned paper as a platform for inexpensive, low-volume, portable bioassays. Angew Chem Int Ed. 2007;3.
Flores-Hernandez DR, Santamaria-Garcia VJ, Melchor-Martínez EM, Sosa-Hernández JE, Parra-Saldívar R, Bonilla-Rios J. Paper and Other Fibrous Materials—A Complete Platform for Biosensing Applications. Biosensors. 2021;11:128.
Dungchai W, Chailapakul O, Henry CS. Electrochemical detection for paper-based microfluidics. Anal Chem. 2009;81:5821–6.
del Valle M. Sensors as green tools in analytical chemistry. Curr Opin Green Sustain Chem. 2021;31:100501.
Kuswandi B, Ensafi AA. Perspective—Paper-Based Biosensors: Trending Topic in Clinical Diagnostics Developments and Commercialization. J Electrochem Soc. 2020;167:037509.
Ahmed S, Bui M-PN, Abbas A. Paper-based chemical and biological sensors: engineering aspects. Biosens Bioelectron. 2016;77:249–63.
Grazioli C, Faura G, Dossi N, Toniolo R, Tubaro F, Terzi F, et al. A colorimetric paper-based smart label soaked with a deep-eutectic solvent for the detection of malondialdehyde. Sens Actuators B Chem. 2021;329:129174.
Toniolo R, Dossi N, Giannilivigni E, Fattori A, Svigelj R, Bontempelli G, et al. Modified screen printed electrode suitable for electrochemical measurements in gas phase. Anal Chem. 2020;92:3689–96.
Toniolo R, Dossi N, Svigelj R, Susmel S, Casella IG, Bontempelli G. Amperometric sniffer for volatile amines based on paper-supported room temperature ionic liquids enabling rapid assessment of fish spoilage. Electroanalysis. 2014;26:1966–74.
Zhang X, Zhi H, Zhu M, Wang F, Meng H, Feng L. Electrochemical/visualdual-readout aptasensor for Ochratoxin A detection integrated into a miniaturized paper-based analytical device. Biosens Bioelectron. 2021;180:113146.
Medina-Sánchez M, Cadevall M, Ros J, Merkoçi A. Eco-friendly electrochemical lab-on-paper for heavy metal detection. Anal Bioanal Chem. 2015;407:8445–9.
Cinti S. Novel paper-based electroanalytical tools for food surveillance. Anal Bioanal Chem. 2019;411:4303–11.
Scherf KA, Uhlig S, Simon K, Frost K, Koehler P, Weiss T, et al. Validation of a qualitative R5 dip-stick for gluten detection with a new mathematical-statistical approach. Qual Assur Saf Crops Foods. 2016;8:309–18.
Wieser H, Koehler P. The biochemical basis of celiac disease. Cereal Chem J. 2008;85:1–13.
Kieffer R, Schurer F, Köhler P, Wieser H. Effect of hydrostatic pressure and temperature on the chemical and functional properties of wheat gluten: studies on gluten, gliadin and glutenin. J Cereal Sci. 2007;45:285–92.
Scherf KA, Poms RE. Recent developments in analytical methods for tracing gluten. J Cereal Sci. 2016;67:112–22.
García E, Llorente M, Hernando A, Kieffer R, Wieser H, Méndez E. Development of a general procedure for complete extraction of gliadins for heat processed and unheated foods. Eur J Gastroenterol Hepatol. 2005;17:529–39.
Doña VV, Fossati CA, Chirdo FG. Interference of denaturing and reducing agents on the antigen/antibody interaction. Impact on the performance of quantitative immunoassays in gliadin analysis. Eur Food Res Technol. 2008;226:591–602.
Xhaferaj M, Alves TO, Ferreira MSL, Scherf KA. Recent progress in analytical method development to ensure the safety of gluten-free foods for celiac disease patients. J Cereal Sci. 2020;96:103114.
Svigelj R, Bortolomeazzi R, Dossi N, Giacomino A, Bontempelli G, Toniolo R. An effective gluten extraction method exploiting pure choline chloride-based deep eutectic solvents (ChCl-DESs). Food Anal Methods. 2017;10:4079–85.
Abbott AP, Boothby D, Capper G, Davies DL, Rasheed RK. Deep eutectic solvents formed between choline chloride and carboxylic acids: versatile alternatives to ionic liquids. J Am Chem Soc. 2004;126:9142–7.
Hansen BB, Spittle S, Chen B, Poe D, Zhang Y, Klein JM, et al. Deep Eutectic Solvents: A Review of Fundamentals and Applications. Chem Rev 2020; acs.chemrev.0c00385.
Zhang Q, De Oliveira VK, Royer S, Jerome F. Deep eutectic solvents: syntheses, properties and applications. Chem Soc Rev. 2012;41:7108–46.
Vian M, Breil C, Vernes L, Chaabani E, Chemat F. Green solvents for sample preparation in analytical chemistry. Curr Opin Green Sustain Chem. 2017;5:44–8.
Yu D, Xue Z, Mu T. Eutectics: formation, properties, and applications. Chem Soc Rev. 2021. https://doi.org/10.1039/D1CS00404B.
Rodríguez-Ramos R, Santana-Mayor Á, Socas-Rodríguez B, Rodríguez-Delgado MÁ. Recent applications of deep eutectic solvents in environmental analysis. Appl Sci. 2021;11:4779.
Svigelj R, Dossi N, Grazioli C, Toniolo R. Deep eutectic solvents (DESs) and their application in biosensor development. Sensors. 2021;21:4263.
Bi W, Tian M, Row KH. Evaluation of alcohol-based deep eutectic solvent in extraction and determination of flavonoids with response surface methodology optimization. J Chromatogr A. 2013;1285:22–30.
Duan L, Dou L-L, Guo L, Li P, Liu E-H. Comprehensive evaluation of deep eutectic solvents in extraction of bioactive natural products. ACS Sustain Chem Eng. 2016;4:2405–11.
Dai Y, Rozema E, Verpoorte R, Choi YH. Application of natural deep eutectic solvents to the extraction of anthocyanins from Catharanthus roseus with high extractability and stability replacing conventional organic solvents. J Chromatogr A. 2016;1434:50–6.
Liu P, Hao J-W, Mo L-P, Zhang Z-H. Recent advances in the application of deep eutectic solvents as sustainable media as well as catalysts in organic reactions. RSC Adv. 2015;5:48675–704.
Toniolo R, Dossi N, Svigelj R, Pigani L, Terzi F, Abollino O, et al. A deep eutectic solvent-based Amperometric sensor for the detection of low oxygen contents in gaseous atmospheres. Electroanalysis. 2016;28:757–63.
Gotor-Fernández V, Paul CE. Deep eutectic solvents for redox biocatalysis. J Biotechnol. 2019;293:24–35.
Seo HB, Gu MB. Aptamer-based sandwich-type biosensors. J Biol Eng. 2017;11:11.
Kang Y, Feng K-J, Chen J-W, Jiang J-H, Shen G-L, Yu R-Q. Electrochemical detection of thrombin by sandwich approach using antibody and aptamer. Bioelectrochemistry. 2008;73:76–81.
Slot IDB, van der Fels-Klerx HJ, Bremer MGEG, Hamer RJ. Immunochemical detection methods for gluten in food products: where do we go from Here? Crit Rev Food Sci Nutr. 2016;56:2455–66.
Svigelj R, Dossi N, Pizzolato S, Toniolo R, Miranda-Castro R, de-los-Santos-Álvarez N, et al. Truncated aptamers as selective receptors in a gluten sensor supporting direct measurement in a deep eutectic solvent. Biosens Bioelectron. 2020;165:112339.
Dhiman A, Anand A, Malhotra A, Khan E, Santra V, Kumar A, et al. Rational truncation of aptamer for cross-species application to detect krait envenomation. Sci Rep. 2018;8:17795.
Amaya-González S, de-los-Santos-Álvarez N, Miranda-Ordieres AJ, Lobo-Castañón MJ. Aptamer binding to celiac disease-triggering hydrophobic proteins: a sensitive gluten detection approach. Anal Chem. 2014;86:2733–9.
Svigelj R, Dossi N, Toniolo R, Miranda-Castro R, de-los-Santos-Álvarez N, Lobo-Castañón MJ. Selection of anti-gluten DNA aptamers in a deep eutectic solvent. Angew Chem. 2018;130:13032–6.
Cao R, Guan L, Li M, Tian J, Shen W. A zero-step functionalization on paper-based biosensing platform for covalent biomolecule immobilization. Sens Bio-Sens Res. 2015;6:13–8.
Malvano F, Albanese D, Pilloton R, Matteo MD. A new label-free impedimetric aptasensor for gluten detection. Food Control. 2017;79:200–6.
Bottari F, Moretto LM, Ugo P. Impedimetric sensing of the immuno-enzymatic reaction of gliadin with a collagen-modified electrode. Electrochem Commun. 2018;97:51–5.
Yu JM, Lee JH, Park J-D, Choi Y-S, Sung J-M, Jang HW. Analyzing Gluten Content in Various Food Products Using Different Types of ELISA Test Kits. Food. 2021;10:108.
Rzychon M, Brohée M, Cordeiro F, Haraszi R, Ulberth F, O’Connor G. The feasibility of harmonizing gluten ELISA measurements. Food Chem. 2017;234:144–54.
Funding
Open access funding provided by Università degli Studi di Udine within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Contributions
Rossella Svigelj: Conceptualization, Investigation, Methodology, Writing—original draft, Visualization. Nicolò Dossi: Writing—review & editing, Visualization. Cristian Grazioli: Writing—review & editing, Visualization. Rosanna Toniolo: Supervision, Conceptualization, Methodology, Writing—review & editing.
Corresponding authors
Ethics declarations
Conflicts of Interest
The authors declare no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Svigelj, R., Dossi, N., Grazioli, C. et al. Paper-based aptamer-antibody biosensor for gluten detection in a deep eutectic solvent (DES). Anal Bioanal Chem 414, 3341–3348 (2022). https://doi.org/10.1007/s00216-021-03653-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-021-03653-5