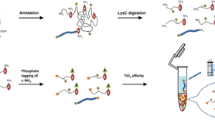
Abstract
Because of structural flexibility and acid lability, the identification of phosphorylated lysine (pLys) peptides is a great challenge. We report here a cleavable hydrophobic derivatization (CHD) strategy for the enrichment and identification of pLys peptides. First, 2,5-dioxopyrrolidin-1-yl-3-(decyldisulfanyl)propanoate was synthesized to react with dephosphorylated lysine peptides, and then the derived peptides were captured by a C18 column, followed by cleavage of the hydrophobic chain, with the specific label left on the target peptides for further identification. By CHD, the enrichment of pLys peptides from interfering peptides (1:1000 mass ratio) was achieved. Furthermore, CHD was applied to screen the pLys targets from Escherichia coli lysates, and 39 pLys sites from 35 proteins were identified. Gene Ontology (GO) analysis showed that these proteins played vital roles in catabolism, metabolism, biogenesis, and biosynthetic processes. All these results demonstrate that CHD might pave the way for comprehensive profiling of the pLys proteome.




Similar content being viewed by others
References
Murat S, Bigot M, Chapron J, Konig GM, Kostenis E, Battaglia G, et al. 5-HT2A receptor-dependent phosphorylation of mGlu2 receptor at serine 843 promotes mGlu2 receptor-operated Gi/o signaling. Mol Psychiatry. 2018. https://doi.org/10.1038/s41380-018-0069-6.
Fuhs SR, Hunter T. pHisphorylation: the emergence of histidine phosphorylation as a reversible regulatory modification. Curr Opin Cell Biol. 2017;45:8–16. https://doi.org/10.1016/j.ceb.2016.12.010.
Besant P, Attwood P, Piggott M. Focus on phosphoarginine and phospholysine. Curr Protein Pept Sci. 2009;10(6):536–50. https://doi.org/10.2174/138920309789630598.
Riley NM, Coon JJ. Phosphoproteomics in the age of rapid and deep proteome profiling. Anal Chem. 2016;88(1):74–94. https://doi.org/10.1021/acs.analchem.5b04123.
Ciesa J, Fraczyk T, Rode W. Phosphorylation of basic amino acid residues in proteins: important but easily missed. Acta Biochim Pol. 2011;58:137–47.
Zetterqvist Ö, Engström L. Isolation of N-ε-[32P]phosphoryl-lysine from rat-liver cell sap after incubation with [32P]adenosine triphosphate. Biochim Biophys Acta. 1967;141(3):523–32. https://doi.org/10.1016/0304-4165(67)90181-x.
Zetterqvist Ö. Further studies on acid-labile [32P]phosphate bound to high-molecular weight material from rat-liver cell sap after incubation with [32P]adenosine triphosphate. Biochim Biophys Acta. 1967;141(3):533–9. https://doi.org/10.1016/0304-4165(67)90182-1.
Wålinder O, Zetterqvist Ö, Engström L. Purification of a bovine liver protein rapidly phosphorylated by adenosine triphosphate. Isolation of 1-32P-phosphohistidine 3-32P-phosphohistidine and N-ε-32P-phospholysine from 32P-labeled protein. J Biol Chem. 1968;243(10):2793–8.
Wålinder O. Identification of a phosphate-incorporating protein from bovine liver as nucleoside diphosphate kinase and isolation of 1-32P-phosphohistidine 3-32P-phosphohistidine and N-ε-32P-phospholysine from erythrocytic nucleoside diphosphate kinase incubated with adenosine triphosphate-32P. J Biol Chem. 1968;243(14):3947–52.
Wei Y, Matthews HR. Identification of phosphohistidine in proteins and purification of protein-histidine kinases. Methods Enzymol. 1991;200:388–414. https://doi.org/10.1016/0076-6879(91)00156-Q.
Ohmori H, Kuba M, Kumon A. Two phosphatases for 6-phospholysine and 3-phosphohistidine from rat brain. J Biol Chem. 1993;268:7625–7.
Hiraishi H, Yokoi F, Kumon A. 3-phosphohistidine and 6-phospholysine are substrates of a 56-kDa inorganic pyrophosphatase from bovine liver. Arch Biochem Biophys. 1998;349(2):381–7. https://doi.org/10.1006/abbi.1997.0480.
Hiraishi H, Yokoi F, Kumon A. Bovine liver phosphoamidase as a protein histidine/lysine phosphatase. J Biochem. 1999;126(2):368–74. https://doi.org/10.1093/oxfordjournals.jbchem.a022459.
Ek P, Ek B, Zetterqvist O. Phosphohistidine phosphatase 1 (PHPT1) also dephosphorylates phospholysine of chemically phosphorylated histone H1 and polylysine. Ups J Med Sci. 2015;120(1):20–7. https://doi.org/10.3109/03009734.2014.996720.
Chen CC, Smith DL, Bruegger BB, Halpern RM, Smith RA. Occurrence and distribution of acid-labile histone phosphates in regenerating rat liver. Biochem. 1974;13:3785–9. https://doi.org/10.1021/bi00715a026.
Chen CC, Bruegger BB, Kern CW, Lin YC, Halpern RM, Smith RA. Phosphorylation of nuclear proteins in rat regenerating liver. Biochem. 1977;16(22):4852–5. https://doi.org/10.1021/bi00641a016.
Bertran-Vicente J, Schümann M, Schmieder P, Krausea E, Hackenberger CPR. Direct access of site-specifically phosphorylated lysine peptides from solid-support. Org Biomol Chem. 2015;13:6839–43. https://doi.org/10.1039/C5OB00734H.
Bertran-Vicente J, Serwa RA, Schumann M, Schmieder P, Krause E, Hackenberger CP. Site-specifically phosphorylated lysine peptides. J Am Chem Soc. 2014;136(39):13622–8. https://doi.org/10.1021/ja507886s.
Bertran-Vicente J, Schumann M, Hackenberger CP, Krause E. Gas-phase rearrangement in lysine phosphorylated peptides during electron-transfer dissociation tandem mass spectrometry. Anal Chem. 2015;87(14):6990–4. https://doi.org/10.1021/acs.analchem.5b01389.
Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJ. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc. 2009;4(4):484–94. https://doi.org/10.1038/nprot.2009.21.
Lin S, Garcia BA. Examining histone posttranslational modification patterns by high-resolution mass spectrometry. Methods Enzymol. 2012;512:3–28. https://doi.org/10.1016/B978-0-12-391940-3.00001-9.
Huesgen PF, Lange PF, Rogers LD, Solis N, Eckhard U, Kleifeld O, et al. LysargiNase mirrors trypsin for protein C-terminal and methylation-site identification. Nat Methods. 2015;12(1):55–8. https://doi.org/10.1038/nmeth.3177.
Acknowledgements
The authors are grateful for financial support from the National Key Research and Development Program of China (2017YFA0505003), the National Natural Science Foundation (91543201, 21505133, 21725506), the Chinese Academy of Sciences Key Project in Frontier Science (QYZDY-SSW-SLH017), and the Innovation Program of Dalian Institute of Chemical Physics, Chinese Academy of Sciences Key (DICP TMSR201601).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Published in the topical collection New Insights into Analytical Science in China with guest editors Lihua Zhang, Hua Cui, and Qiankun Zhuang.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hu, Y., Li, Y., Gao, H. et al. Cleavable hydrophobic derivatization strategy for enrichment and identification of phosphorylated lysine peptides. Anal Bioanal Chem 411, 4159–4166 (2019). https://doi.org/10.1007/s00216-019-01770-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-019-01770-w