Abstract
Here we report a bacteria detection method based on a flow cytometric bead system and 16S rRNA-targeted oligonucleotide probes. Polymerase chain reaction (PCR) was first used to acquire bacterial DNA including bacteria-specific sequences. Half of the resulting target DNA was then captured by a capture probe immobilized on a magnetic microbead (MB) surface. The other half of the target DNA was hybridized with a fluorescence-labeled signal probe. In this manner, a sandwich DNA hybridization involving a MB-based capture probe, the target DNA, and a signal probe was realized. The MB carriers modified with reporter dye were analyzed one by one by flow cytometry through a capillary. Using PCR amplicons and this flow cytometric bead system, a detection limit of 180 cfu mL−1 was achieved, along with high selectivity that permitted the discrimination of different targets when challenged with control bacteria targets and multiplexing capabilities that enabled the simultaneous detection of two kinds of bacteria. Given these advantages, the developed method can be used for the highly sensitive and specific PCR amplicon analysis of DNA extracted from a fresh bacterial culture, as well as multiplex target analysis.

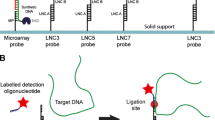
The flow cytometric bead system with 16S rRNA-targeted oligonucleotide probes for bacteria detection developed in this work. This system is highly specific and sensitive, with a detection limit of 180 cfu mL−1 bacteria.





Similar content being viewed by others
References
Ivnitski D, Abdel-Hamid I, Atanasov P, Wilkins E. Biosensors for detection of pathogenic bacteria. Biosens Bioelectron. 1999;14(7):599–624.
Wang Y, Salazar JK. Culture-independent rapid detection methods for bacterial pathogens and toxins in food matrices. Compr Rev Food Sci F. 2016;15(1):183–205.
Yoo SM, Lee SY. Optical biosensors for the detection of pathogenic microorganisms. Trends Biotechnol. 2016;34(1):7–25.
Duvernoy M-C, Mora T, Ardré M, Croquette V, Bensimon D, Quilliet C, et al. Asymmetric adhesion of rod-shaped bacteria controls microcolony morphogenesis. Nat Commun. 2018;9(1):1120.
Russo TA, Johnson JR. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 2003;5(5):449–56.
Nash AA, Dalziel RG, Fitzgerald JR. Mims’ pathogenesis of infectious disease: Cambridge: Academic Press; 2015.
Manoharan R, Ghiamati E, Dalterio R, Britton K, Nelson W, Sperry J. UV resonance Raman spectra of bacteria, bacterial spores, protoplasts and calcium dipicolinate. J Microbiol Meth. 1990;11(1):1–15.
Diehl F, Li M, He Y, Kinzler KW, Vogelstein B, Dressman D. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods. 2006;3(7):551.
Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, et al. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239(4839):487–91.
Zoetendal EG, Akkermans AD, De Vos WM. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microb. 1998;64(10):3854–9.
Duro G, Barbieri R, Ribaudo MR, Feo S, Izzo V. A downward capillary blotting procedure from hundred base pairs to hundred kilobases nucleic acids. Anal Chem. 1995;225(2):360–2.
Yean CY, Kamarudin B, Ozkan DA, Yin LS, Lalitha P, Ismail A, et al. Enzyme-linked amperometric electrochemical genosensor assay for the detection of PCR amplicons on a streptavidin-treated screen-printed carbon electrode. Anal Chem. 2008;80(8):2774–9.
Matsuki T, Watanabe K, Fujimoto J, Takada T, Tanaka R. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl Environ Microbiol. 2004;70(12):7220–8.
Vermes I, Haanen C, Steffens-Nakken H, Reutellingsperger C. A novel assay for apoptosis flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled annexin V. J Immunol Methods. 1995;184(1):39–51.
Lu J, Paulsen IT, Jin D. Application of exonuclease III-aided target recycling in flow cytometry: DNA detection sensitivity enhanced by orders of magnitude. Anal Chem. 2013;85(17):8240–5.
Kounelli ML, Kalogianni DP. A sensitive DNA-based fluorometric method for milk authenticity of dairy products based on spectrally distinct microspheres. Eur Food Res Technol. 2017;243(10):1773–81.
Janczuk M. Richter Ł, Hoser G, Kawiak J, Łoś M, Niedziółka-Jönsson J, et al. Bacteriophage-based bioconjugates as a flow cytometry probe for fast bacteria detection. Bioconjug Chem. 2016;28(2):419–25.
Suárez H, Gámez-Valero A, Reyes R, López-Martín S, Rodríguez MJ, Carrascosa JL, et al. A bead-assisted flow cytometry method for the semi-quantitative analysis of extracellular vesicles. Sci Rep. 2017;7(1):11271.
Volgers C, Benedikter BJ, Grauls GE, Savelkoul PH, Stassen FR. Bead-based flow-cytometry for semi-quantitative analysis of complex membrane vesicle populations released by bacteria and host cells. Microbiol Res. 2017;200:25–32.
Huang P-JJ, Liu J. Flow cytometry-assisted detection of adenosine in serum with an immobilized aptamer sensor. Anal Chem. 2010;82(10):4020–6.
Zeng Y, Zhang D, Qi P, Zheng LB. Colorimetric detection of DNA by using target catalyzed DNA nanostructure assembly and unmodified gold nanoparticles. Microchim Acta. 2017;184(12):4809–15.
Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. PNAS. 1985;82(20):6955–9.
Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer K-H, et al. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol. 2014;12(9):635–45.
Zeng Y, Wan Y, Zhang D. Lysozyme as sensitive reporter for fluorometric and PCR based detection of E. coli and S. aureus using magnetic microbeads. Microchim Acta. 2016;183(2):741–8.
Chung HJ, Castro CM, Im H, Lee H, Weissleder R. A magneto-DNA nanoparticle system for rapid detection and phenotyping of bacteria. Nat Nanotech. 2013;8(5):369–75.
Mariani S, Minunni M. Surface plasmon resonance applications in clinical analysis. Anal Bioanal Chem. 2014;406(9–10):2303–23.
Adkins JA, Boehle K, Friend C, Chamberlain B, Bisha B, Henry CS. Colorimetric and electrochemical bacteria detection using printed paper- and transparency-based analytic devices. Anal Chem. 2017;89(6):3613–21.
Masdor NA, Altintas Z, Tothill IE. Sensitive detection of Campylobacter jejuni using nanoparticles enhanced QCM sensor. Biosens Bioelectron. 2016;78:328–36.
Poller A-M, Spieker E, Lieberzeit PA, Preininger C. Surface imprints: advantageous application of Ready2use materials for bacterial QCM-sensors. ACS Appl Materials Inter. 2016;9(1):1129–35.
Wang L, Wang R, Chen F, Jiang T, Wang H, Slavik M, et al. QCM-based aptamer selection and detection of Salmonella typhimurium. Food Chem. 2017;221:776–82.
Wan Y, Zhu G. Dopamine-mediated immunoassay for bacteria detection. Anal Bioanal Chem. 2017;409(26):6091–6.
Zhang Q, Wang X-D, Tian T, Chu L-Q. Incorporation of multilayered silver nanoparticles into polymer brushes as 3-dimensional SERS substrates and their application for bacteria detection. Appl Surf Sci. 2017;407:185–91.
Maldonado J, González-Guerrero AB, Domínguez C, Lechuga LM. Label-free bimodal waveguide immunosensor for rapid diagnosis of bacterial infections in cirrhotic patients. Biosens Bioelectron. 2016;85:310–6.
Acknowledgements
We gratefully acknowledge the support provided by the National Natural Science Foundation of China (41876101) and project funding from the China Postdoctoral Science Foundation (2016 M602199), the Young Elite Scientists Sponsorship Program by CAST (2018QNRC001), the Applied Basic Research Program of Nantong (GY12017005), and the AoShan Talent Program supported by the Qingdao National Laboratory for Marine Science and Technology.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 414 kb)
Rights and permissions
About this article
Cite this article
Zeng, Y., Zhang, D. & Qi, P. Combination of a flow cytometric bead system with 16S rRNA-targeted oligonucleotide probes for bacteria detection. Anal Bioanal Chem 411, 2161–2168 (2019). https://doi.org/10.1007/s00216-019-01651-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-019-01651-2