Abstract
Urine is an attractive and non-invasive alternative source to tissue, blood or other biofluids for biomarker screening in clinical research. In normal human adult urine, 48% of the total urinary protein is in the sediment, 49% is soluble and the remaining 3% is contained in urinary extracellular vesicles (EVs). The soluble proteins and EV proteins in urine have attracted particular attention in recent years as cancer diagnostics. Furthermore, considering the important role of N-glycoproteins in practically all physiological processes, including regulating receptor-ligand binding, cell-cell interactions, inflammatory response and tumour progression, N-glycoproteome in human urine is an invaluable target for monitoring the physiological status and pathological changes of the kidney and urinary tract. Given the different origins of the soluble proteins and EV proteins in the urine, different N-glycoproteome patterns exist. Therefore, isolating the soluble N-glycoproteins and EV N-glycoproteins for separate analysis will provide a more specific and comprehensive view and provide a deeper understanding of human urinary N-glycoproteome. In this work, we developed a sequential separation method that isolates urinary soluble proteins and EV proteins via stepwise ultrafiltration based on their obvious size difference. A facile and reproducible protein isolation was achieved using this strategy. Subsequent N-glycoproteome enrichment and identification revealed distinct patterns in the two sub-proteomes of urine with more than 60% differential N-glycopeptides. A more comprehensive picture of the urinary N-glycoproteome with close to 1800 identified N-glycopeptides was obtained by this new analysis strategy, therefore making it advantageous for urinary biomarker screening.

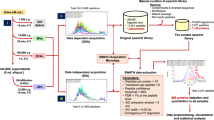
A sequential separation method that isolates urinary soluble proteins and EV proteins via stepwise ultrafiltration was developed in this work. Subsequent N-glycopeptides enrichment and mass spectrometry analysis reveals distinct N-glycoproteome patterns in the two sub-proteomes of urine and a deep mapping of close to 1800 N-glycopeptides.





Similar content being viewed by others
References
Brown N, Segev G, Francey T, Kass P, Cowgill L. Glomerular filtration rate, urine production, and fractional clearance of electrolytes in acute kidney injury in dogs and their association with survival. J Vet Intern Med. 2015;29(1):28–34.
Moon PG, You S, Lee JE, Hwang D, Baek MC. Urinary exosomes and proteomics. Mass Spectrom Rev. 2011;30(6):1185–202.
Zoja C, Abbate M, Remuzzi G. Progression of renal injury toward interstitial inflammation and glomerular sclerosis is dependent on abnormal protein filtration. Nephro Dial Transpl. 2014;30(5):706–12.
Pontillo C, Jacobs L, Staessen JA, Schanstra JP, Rossing P, Heerspink HJ, et al. A urinary proteome-based classifier for the early detection of decline in glomerular filtration. Nephro Dial Transpl. 2016;32(9):1510–6.
Mok CC, Soliman S, Ho LY, Mohamed FA, Mohamed FI, Mohan C. Urinary angiostatin, CXCL4 and VCAM-1 as biomarkers of lupus nephritis. Arthritis Res Ther. 2018;20(1):6–16.
Leyten GH, Hessels D, Jannink SA, Smit FP, de Jong H, Cornel EB, et al. Prospective multicentre evaluation of PCA3 and TMPRSS2-ERG gene fusions as diagnostic and prognostic urinary biomarkers for prostate cancer. Eur Urol. 2014;65(3):534–42.
Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101(36):13368–73.
Gámez-Valero A, Lozano-Ramos SI, Bancu I, Lauzurica-Valdemoros R, Borràs FE. Urinary extracellular vesicles as source of biomarkers in kidney diseases. Front Immunol. 2015;6(6):1–10.
Dear JW, Street JM, Bailey MA. Urinary exosomes: a reservoir for biomarker discovery and potential mediators of intrarenal signalling. Proteomics. 2013;13(10–11):1572–80.
Zhang Y, Wang X-F. A niche role for cancer exosomes in metastasis. Nat Cell Biol. 2015;17(6):709–11.
Buzas EI, György B, Nagy G, Falus A, Gay S. Emerging role of extracellular vesicles in inflammatory diseases. Nat Rev Rheumatol. 2014;10(6):356–64.
Huang-Doran I, Zhang C-Y, Vidal-Puig A. Extracellular vesicles: novel mediators of cell communication in metabolic disease. Trends Endocrinol Metab. 2017;28(1):3–18.
Chen I-H, Aguilar HA, Paez Paez JS, Wu X, Pan L, Wendt MK, et al. An analytical pipeline for discovery and verification of glycoproteins from plasma-derived extracellular vesicles as breast cancer biomarkers. Anal Chem. 2018;90(10):6307–13.
Yang Q, Wang L-X. Chemoenzymatic glycan remodeling of natural and recombinant glycoproteins. Methods Enzymol. 2017;597(1):265–81.
Chen Y, Xiong Z, Zhang L, Zhao J, Zhang Q, Peng L, et al. Facile synthesis of zwitterionic polymer-coated core–shell magnetic nanoparticles for highly specific capture of N-linked glycopeptides. Nanoscale. 2015;7(7):3100–8.
Jiang B, Liang Y, Wu Q, Jiang H, Yang K, Zhang L, et al. New GO–PEI–Au–L-Cys ZIC-HILIC composites: synthesis and selective enrichment of glycopeptides. Nanoscale. 2014;6(11):5616–9.
Hang Q, Isaji T, Hou S, Wang Y, Fukuda T, Gu J. A key regulator of cell adhesion: identification and characterization of important N-glycosylation sites on integrin α5 for cell migration. Mol Cell Biol. 2017;37(9):1–14.
Zhuo Y, Yang J-Y, Moremen KW, Prestegard JH. Glycosylation alters dimerization properties of a cell-surface signaling protein, carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1). J Biol Chem. 2016;291(38):20085–95.
Zhao D, Liang L, Wang S, Nakao T, Li Y, Liu L, et al. Glycosylation of the hemagglutinin protein of H5N1 influenza virus increases its virulence in mice by exacerbating the host immune response. J Virol. 2017;91(7):1–14.
Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15(9):540–55.
Stowell SR, Ju T, Cummings RD. Protein glycosylation in cancer. Annu Rev Pathol. 2015;10(1):473–510.
Guo Z, Liu X, Li M, Shao C, Tao J, Sun W, et al. Differential urinary glycoproteome analysis of type 2 diabetic nephropathy using 2D-LC–MS/MS and iTRAQ quantification. J Transl Med. 2015;13(1):371–88.
Saraswat M, Joenväära S, Musante L, Peltoniemi H, Holthofer H. Renkonen R. N-linked (N-) glycoproteomics of urimary exosomes. Mol Cell Proteomics. 2015;14(2):263–76.
Pan L, Aguilar HA, Wang L, Iliuk A, Tao WA. Three-dimensionally functionalized reverse phase glycoprotein array for cancer biomarker discovery and validation. J Am Chem Soc. 2016;138(47):15311–4.
Jia X, Chen J, Sun S, Yang W, Yang S, Shah P, et al. Detection of aggressive prostate cancer associated glycoproteins in urine using glycoproteomics and mass spectrometry. Proteomics. 2016;16(23):2989–96.
Wang L, Li F, Sun W, Wu S, Wang X, Zhang L, et al. Concanavalin A-captured glycoproteins in healthy human urine. Mol Cell Proteomics. 2006;5(3):560–2.
Saraswat M, Joenväära S, Musante L, Peltoniemi H, Holthofer H. Renkonen R. N-linked (N-) glycoproteomics of urinary exosomes. Mol Cell Proteomics. 2015;14(2):263–76.
Pisitkun T, Johnstone R, Knepper MA. Discovery of urinary biomarkers. Mol Cell Proteomics. 2006;5(10):1760–71.
Sharma K, Schmitt S, Bergner CG, Tyanova S, Kannaiyan N, Manrique-Hoyos N, et al. Cell type–and brain region–resolved mouse brain proteome. Nat Neurosci. 2015;18(12):1819–31.
Doll S, Dreßen M, Geyer PE, Itzhak DN, Braun C, Doppler SA, et al. Region and cell-type resolved quantitative proteomic map of the human heart. Nat Commun. 2017;8(1):1469–82.
Santa C, Anjo SI, Manadas B. Protein precipitation of diluted samples in SDS-containing buffer with acetone leads to higher protein recovery and reproducibility in comparison with TCA/acetone approach. Proteomics. 2016;16(13):1847–51.
Livshits MA, Khomyakova E, Evtushenko EG, Lazarev VN, Kulemin NA, Semina SE, et al. Isolation of exosomes by differential centrifugation: theoretical analysis of a commonly used protocol. Sci Rep. 2015;5(1):17319–32.
Salih M, Zietse R, Hoorn EJ. Urinary extracellular vesicles and the kidney: biomarkers and beyond. Am J Physiol Renal Physiol. 2014;306(11):F1251–9.
Motamedinia P, Scott AN, Bate KL, Sadeghi N, Salazar G, Shapiro E, et al. Urine exosomes for non-invasive assessment of gene expression and mutations of prostate cancer. PLoS One. 2016;11(5):1–15.
Liang L-G, Kong M-Q, Zhou S, Sheng Y-F, Wang P, Yu T, et al. An integrated double-filtration microfluidic device for isolation, enrichment and quantification of urinary extracellular vesicles for detection of bladder cancer. Sci Rep. 2017;7(1):46224–34.
Cheruvanky A, Zhou H, Pisitkun T, Kopp JB, Knepper MA, Yuen PS, et al. Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Am J Physiol Renal Physiol. 2007;292(5):F1657–61.
Pan Y, Ma C, Tong W, Fan C, Zhang Q, Zhang W, et al. Preparation of sequence-controlled triblock copolymer-grafted silica microparticles by sequential-ATRP for highly efficient glycopeptides enrichment. Anal Chem. 2014;87(1):656–62.
Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359–62.
Wu M, Ouyang Y, Wang Z, Zhang R, Huang P-H, Chen C, et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc Natl Acad Sci U S A. 2017;114(40):10584–9.
Merchant ML, Rood IM, Deegens JK, Klein JB. Isolation and characterization of urinary extracellular vesicles: implications for biomarker discovery. Nat Rev Nephrol. 2017;13(12):731–49.
Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2008;37(1):1–13.
Hu S, Musante L, Tataruch D, Xu X, Kretz O, Henry M, et al. Purification and identification of membrane proteins from urinary extracellular vesicles using triton X-114 phase partitioning. J Proteome Res. 2017;17(1):86–96.
Acknowledgments
This study was supported by the National Key Program for Basic Research of China (No. 2017YFC0906703, 2016YFA0501403, 2018YFF0212505, 2017YFA0505002), National Natural Science Foundation of China (No. 21675172), National Key Laboratory of Proteomics Grant (SKLP-K201706), BPRC-Tianjin Baodi Hospital Joint Center Grant (TMRC2014Z03), Innovation Project (16CXZ027) and Beijing Science and Technology Plan Project (No. Z161100002616036).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Live subject statement
The authors state that all experiments were performed in compliance with the relevant laws and institutional guidelines. The institutional committee of the National Centre for Protein Science Beijing has approved the experiments with live subjects. The authors also state that informed consent was obtained for any experimentation with human subjects and National Centre for Protein Science Beijing is committed to the protection and safety of human subjects involved in research.
Rights and permissions
About this article
Cite this article
Huo, B., Chen, M., Chen, J. et al. A sequential separation strategy for facile isolation and comprehensive analysis of human urine N-glycoproteome. Anal Bioanal Chem 410, 7305–7312 (2018). https://doi.org/10.1007/s00216-018-1338-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-018-1338-6