Abstract
Brain extracellular matrix (ECM) is a highly organized system that consists of collagens, noncollagenous proteins, glycoproteins, hyaluronan, and proteoglycans. Recognized physiological roles of ECM include developmental regulation, tissue homeostasis, cell migration, cell proliferation, cell differentiation, neuronal plasticity, and neurite outgrowth. Aberrant ECM structure is associated with brain neurodegenerative conditions. This review focuses on two neurodegenerative conditions, schizophrenia and Alzheimer’s disease, and summarizes recent findings of altered ECM components, including proteoglycans, glycosaminoglycans, proteins, and glycoproteins, and proteins and genes related to other brain components. The scope includes immunohistochemical, genomics, transcriptomics, proteomics, and glycomics studies, and a critical assessment of current state of proteomic studies for neurodegenerative disorders. The intent is to summarize the ECM molecular alterations associated with neurodegenerative pathophysiology.

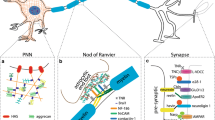
Brain extracellular matrix showing HSPGs, CSPGs, HA, collagens, and other glycoproteins.


Similar content being viewed by others
Abbreviations
- Aβ:
-
Amyloid beta
- AD:
-
Alzheimer’s disease
- ADAM:
-
A disintegrin and metalloprotease
- ADAMTS:
-
A disintegrin and metalloprotease with thrombospondin motifs
- APP:
-
Amyloid beta A4 precursor protein
- BACE1:
-
β-site amyloid beta A4 precursor protein cleaving enzyme 1
- CS:
-
Chondroitin sulfate
- CSF:
-
Cerebrospinal fluid
- CSPG:
-
Chondroitin sulfate proteoglycan
- DS:
-
Dermatan sulfate
- ECM:
-
Extracellular matrix
- EGF:
-
Epidermal growth factor
- GAG:
-
Glycosaminoglycan
- GalNAc:
-
N-acetylgalactosamine
- GAPDH:
-
Glyceraldehyde 3-phosphate dehydrogenase
- GlcNAc:
-
N-acetylglucosamine
- GlcNAcT:
-
N-acetylglucosamine transferase
- HA:
-
Hyaluronan
- HS:
-
Heparan sulfate
- HSPG:
-
Heparan sulfate proteoglycan
- KS:
-
Keratan sulfate
- LC:
-
Liquid chromatography
- MMP:
-
Matrix metalloprotease
- MS/MS:
-
Tandem mass spectrometry
- PFC:
-
Prefrontal cortex
- PG:
-
Proteoglycan
- PNN:
-
Perineuronal net
- PTM:
-
Post-translational modification
- RPTPβ:
-
Receptor phosphotyrosine phosphatase beta/zeta
- SZ:
-
Schizophrenia
- WFA:
-
Wisteria floribunda agglutinin
References
Nicholson C, Sykova E. Extracellular space structure revealed by diffusion analysis. Trends Neurosci. 1998;21:207–15.
Novak U, Kaye AH. Extracellular matrix and the brain: components and function. J Clin Neurosci. 2000;7:280–90.
Smith PD, Coulson-Thomas VJ, Foscarin S, Kwok JC, Fawcett JW. “GAG-ing with the neuron”: the role of glycosaminoglycan patterning in the central nervous system. Exp Neurol. 2015;274:100–14.
Bonneh-Barkay D, Wiley CA. Brain extracellular matrix in neurodegeneration. Brain Pathol. 2009;19:573–85.
Berretta S. Extracellular matrix abnormalities in schizophrenia. Neuropharmacology. 2012;62:1584–97.
Busch SA, Silver J. The role of extracellular matrix in CNS regeneration. Curr Opin Neurobiol. 2007;17:120–7.
Green REA. Editorial on brain injury as a neurodegenerative disorder. Front Hum Neurosci. 2016. doi:10.3389/fnhum.2015.00615.
Kjellen L, Lindahl U. Proteoglycans: structures and interactions. Annu Rev Biochem. 1991;60:443–75.
Iozzo RV, Schaefer L. Proteoglycan form and function: a comprehensive nomenclature of proteoglycans. Matrix Biol. 2015;42:11–55.
Schaefer L, Iozzo RV. Biological functions of the small leucine-rich proteoglycans: from genetics to signal transduction. J Biol Chem. 2008;283:21305–9.
Yamaguchi Y. Lecticans: organizers of the brain extracellular matrix. Cell Mol Life Sci. 2000;57:276–89.
Iozzo RV. Basement membrane proteoglycans: from cellar to ceiling. Nat Rev Mol Cell Biol. 2005;6:646–56.
Lord MS, Chuang CY, Melrose J, Davies MJ, Iozzo RV, Whitelock JM. The role of vascular-derived perlecan in modulating cell adhesion, proliferation and growth factor signaling. Matrix Biol. 2014;35:112–22.
Wilusz RE, Sanchez-Adams J, Guilak F. The structure and function of the pericellular matrix of articular cartilage. Matrix Biol. 2014;39:25–32.
De Rossi G, Evans AR, Kay E, Woodfin A, McKay TR, Nourshargh S, et al. Shed syndecan-2 inhibits angiogenesis. J Cell Sci. 2014;127:4788–99.
Filmus J, Capurro M, Rast J. Glypicans. Genome Biol. 2008;9:224.
Yang BL, Zhang Y, Cao L, Yang BB. Cell adhesion and proliferation mediated through the G1 domain of versican. J Cell Biochem. 1999;72:210–20.
Olin AI, Morgelin M, Sasaki T, Timpl R, Heinegard D, Aspberg A. The proteoglycans aggrecan and versican form networks with fibulin-2 through their lectin domain binding. J Biol Chem. 2001;276:1253–61.
Zhang Y, Cao L, Yang BL, Yang BB. The G3 domain of versican enhances cell proliferation via epidermal growth factor-like motifs. J Biol Chem. 1998;273:21342–51.
Beller JA, Snow DM. Proteoglycans: road signs for neurite outgrowth. Neural Regen Res. 2014;9:343–55.
Friedlander DR, Milev P, Karthikeyan L, Margolis RK, Margolis RU, Grumet M. The neuronal chondroitin sulfate proteoglycan neurocan binds to the neural cell adhesion molecules Ng-CAM/L1/NILE and N-CAM, and inhibits neuronal adhesion and neurite outgrowth. J Cell Biol. 1994;125:669–80.
Yamada H, Fredette B, Shitara K, Hagihara K, Miura R, Ranscht B, et al. The brain chondroitin sulfate proteoglycan brevican associates with astrocytes ensheathing cerebellar glomeruli and inhibits neurite outgrowth from granule neurons. J Neurosci. 1997;17:7784–95.
Snow DM, Lemmon V, Carrino DA, Caplan AI, Silver J. Sulfated proteoglycans in astroglial barriers inhibit neurite outgrowth in vitro. Exp Neurol. 1990;109:111–30.
Dauth S, Grevesse T, Pantazopoulos H, Campbell PH, Maoz BM, Berretta S, et al. Extracellular matrix protein expression is brain region dependent. J Comp Neurol. 2016;524:1309–36.
Kwok JC, Dick G, Wang D, Fawcett JW. Extracellular matrix and perineuronal nets in CNS repair. Dev Neurobiol. 2011;71:1073–89.
Zoltan-Jones A, Huang L, Ghatak S, Toole BP. Elevated hyaluronan production induces mesenchymal and transformed properties in epithelial cells. J Biol Chem. 2003;278:45801–10.
Spicer AP, Tien JY. Hyaluronan and morphogenesis. Birth Defects Res C Embryo Today. 2004;72:89–108.
McAtee CO, Barycki JJ, Simpson MA. Emerging roles for hyaluronidase in cancer metastasis and therapy. Adv Cancer Res. 2014;123:1–34.
Margolis RK, Margolis RU. Nervous tissue proteoglycans. EXS. 1993;49:429–46.
Schlessinger J, Lax I, Lemmon M. Regulation of growth factor activation by proteoglycans: what is the role of the low affinity receptors? Cell. 1995;83:357–60.
Neufeld G, Cohen T, Gengrinovitch S, Poltorak Z. Vascular endothelial growth factor (VEGF) and its receptors. FASEB J. 1999;13:9–22.
Milev P, Monnerie H, Popp S, Margolis RK, Margolis RU. The core protein of the chondroitin sulfate proteoglycan phosphacan is a high-affinity ligand of fibroblast growth factor-2 and potentiates its mitogenic activity. J Biol Chem. 1998;273:21439–42.
Celio MR, Spreafico R, De Biasi S, Vitellaro-Zuccarello L. Perineuronal nets: past and present. Trends Neurosci. 1998;21:510–5.
Pyka M, Wetzel C, Aguado A, Geissler M, Hatt H, Faissner A. Chondroitin sulfate proteoglycans regulate astrocyte-dependent synaptogenesis and modulate synaptic activity in primary embryonic hippocampal neurons. Eur J Neurosci. 2011;33:2187–202.
Reimers S, Hartlage-Rubsamen M, Bruckner G, Rossner S. Formation of perineuronal nets in organotypic mouse brain slice cultures is independent of neuronal glutamatergic activity. Eur J Neurosci. 2007;25:2640–8.
Andersen SL. Trajectories of brain development: point of vulnerability or window of opportunity? Neurosci Biobehav Rev. 2003;27:3–18.
Woo TU. Neurobiology of schizophrenia onset. Curr Top Behav Neurosci. 2014;16:267–95.
McGorry PD. Early intervention in psychotic disorders: beyond debate to solving problems. Br J Psychiatry Suppl. 2005;48:108–10.
McGlashan TH, Hoffman RE. Schizophrenia as a disorder of developmentally reduced synaptic connectivity. Arch Gen Psychiatry. 2000;57:637–48.
Takahashi N, Sakurai T, Bozdagi-Gunal O, Dorr NP, Moy J, Krug L, et al. Increased expression of receptor phosphotyrosine phosphatase-beta/zeta is associated with molecular, cellular, behavioral and cognitive schizophrenia phenotypes. Transl Psychiatry. 2011. doi:10.1038/tp.2011.8.
Pantazopoulos H, Woo TU, Lim MP, Lange N, Berretta S. Extracellular matrix-glial abnormalities in the amygdala and entorhinal cortex of subjects diagnosed with schizophrenia. Arch Gen Psychiatry. 2010;67:155–66.
Mauney SA, Athanas KM, Pantazopoulos H, Shaskan N, Passeri E, Berretta S, et al. Developmental pattern of perineuronal nets in the human prefrontal cortex and their deficit in schizophrenia. Biol Psychiatry. 2013;74:427–35.
Shah A, Lodge DJ. A loss of hippocampal perineuronal nets produces deficits in dopamine system function: relevance to the positive symptoms of schizophrenia. Transl Psychiatry. 2013. doi:10.1038/tp.2012.145.
Pantazopoulos H, Berretta S. In sickness and in health: perineuronal nets and synaptic plasticity in psychiatric disorders. Neural Plast. 2016. doi:10.1155/2016/9847696.
Habl G, Schmitt A, Zink M, von Wilmsdorff M, Yeganeh-Doost P, Jatzko A, et al. Decreased reelin expression in the left prefrontal cortex (BA9) in chronic schizophrenia patients. Neuropsychobiology. 2012;66:57–62.
Enwright JF, Sanapala S, Foglio A, Berry R, Fish KN, Lewis DA. Reduced labeling of parvalbumin neurons and perineuronal nets in the dorsolateral prefrontal cortex of subjects with schizophrenia. Neuropsychopharmacology. 2016. doi:10.1038/npp.2016.24.
Pietersen CY, Mauney SA, Kim SS, Lim MP, Rooney RJ, Goldstein JM, et al. Molecular profiles of pyramidal neurons in the superior temporal cortex in schizophrenia. J Neurogenet. 2014;28:53–69.
Impagnatiello F, Guidotti AR, Pesold C, Dwivedi Y, Caruncho H, Pisu MG, et al. A decrease of reelin expression as a putative vulnerability factor in schizophrenia. Proc Natl Acad Sci U S A. 1998;95:15718–23.
Guidotti A, Auta J, Davis JM, Di-Giorgi-Gerevini V, Dwivedi Y, Grayson DR, et al. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch Gen Psychiatry. 2000;57:1061–9.
Eastwood SL, Harrison PJ. Cellular basis of reduced cortical reelin expression in schizophrenia. Am J Psychiatry. 2006;163:540–2.
Prabakaran S, Swatton JE, Ryan MM, Huffaker SJ, Huang JT, Griffin JL, et al. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry. 2004;9:684–97. 643.
Mahadik SP, Mukherjee S, Wakade CG, Laev H, Reddy RR, Schnur DB. Decreased adhesiveness and altered cellular distribution of fibronectin in fibroblasts from schizophrenic patients. Psychiatry Res. 1994;53:87–97.
Miyamae Y, Nakamura Y, Kashiwagi Y, Tanaka T, Kudo T, Takeda M. Altered adhesion efficiency and fibronectin content in fibroblasts from schizophrenic patients. Psychiatry Clin Neurosci. 1998;52:345–52.
Walsh MT, Ryan M, Hillmann A, Condren R, Kenny D, Dinan T, et al. Elevated expression of integrin αIIb βIIIa in drug-naive, first-episode schizophrenic patients. Biol Psychiatry. 2002;52:874–9.
Domenici E, Wille DR, Tozzi F, Prokopenko I, Miller S, McKeown A, et al. Plasma protein biomarkers for depression and schizophrenia by multi analyte profiling of case-control collections. PLoS One. 2010. doi:10.1371/journal.pone.0009166.
Yamamori H, Hashimoto R, Ishima T, Kishi F, Yasuda Y, Ohi K, et al. Plasma levels of mature brain-derived neurotrophic factor (BDNF) and matrix metalloproteinase-9 (MMP-9) in treatment-resistant schizophrenia treated with clozapine. Neurosci Lett. 2013;556:37–41.
Shibasaki C, Takebayashi M, Itagaki K, Abe H, Kajitani N, Okada-Tsuchioka M, et al. Altered serum levels of matrix metalloproteinase-2, -9 in response to electroconvulsive therapy for mood disorders. Int J Neuropsychopharmacol. 2016. doi:10.1093/ijnp/pyw019.
Buxbaum JD, Georgieva L, Young JJ, Plescia C, Kajiwara Y, Jiang Y, et al. Molecular dissection of NRG1-ERBB4 signaling implicates PTPRZ1 as a potential schizophrenia susceptibility gene. Mol Psychiatry. 2008;13:162–72.
Parlapani E, Schmitt A, Wirths O, Bauer M, Sommer C, Rueb U, et al. Gene expression of neuregulin-1 isoforms in different brain regions of elderly schizophrenia patients. World J Biol Psychiatry. 2010;11:243–50.
Keri S, Beniczky S, Kelemen O. Suppression of the P50 evoked response and neuregulin 1-induced AKT phosphorylation in first-episode schizophrenia. Am J Psychiatry. 2010;67:444–50.
Law AJ, Kleinman JE, Weinberger DR, Weickert CS. Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum Mol Genet. 2007;16:129–41.
Saia-Cereda VM, Cassoli JS, Schmitt A, Falkai P, Nascimento JM, Martins-de-Souza D. Proteomics of the corpus callosum unravel pivotal players in the dysfunction of cell signaling, structure, and myelination in schizophrenia brains. Eur Arch Psychiatry Clin Neurosci. 2015;265:601–12.
Martins-de-Souza D, Maccarrone G, Wobrock T, Zerr I, Gormanns P, Reckow S, et al. Proteome analysis of the thalamus and cerebrospinal fluid reveals glycolysis dysfunction and potential biomarkers candidates for schizophrenia. J Psychiatr Res. 2010;44:1176–89.
English JA, Dicker P, Focking M, Dunn MJ, Cotter DR. 2-D DIGE analysis implicates cytoskeletal abnormalities in psychiatric disease. Proteomics. 2009;9:3368–82.
Focking M, Dicker P, English JA, Schubert KO, Dunn MJ, Cotter DR. Common proteomic changes in the hippocampus in schizophrenia and bipolar disorder and particular evidence for involvement of cornu ammonis regions 2 and 3. Arch Gen Psychiatry. 2011;68:477–88.
Schubert KO, Focking M, Cotter DR. Proteomic pathway analysis of the hippocampus in schizophrenia and bipolar affective disorder implicates 14-3-3 signaling, aryl hydrocarbon receptor signaling, and glucose metabolism: potential roles in GABAergic interneuron pathology. Schizophr Res. 2015;167:64–72.
Martins-de-Souza D, Gattaz WF, Schmitt A, Maccarrone G, Hunyadi-Gulyas E, Eberlin MN, et al. Proteomic analysis of dorsolateral prefrontal cortex indicates the involvement of cytoskeleton, oligodendrocyte, energy metabolism and new potential markers in schizophrenia. J Psychiatr Res. 2009;43:978–86.
Sivagnanasundaram S, Crossett B, Dedova I, Cordwell S, Matsumoto I. Abnormal pathways in the genu of the corpus callosum in schizophrenia pathogenesis: a proteome study. Proteomics Clin Appl. 2007;1:1291–305.
Steiner J, Schmitt A, Schroeter ML, Bogerts B, Falkai P, Turck CW, et al. S100B is downregulated in the nuclear proteome of schizophrenia corpus callosum. Eur Arch Psychiatry Clin Neurosci. 2014;264:311–6.
Stanta JL, Saldova R, Struwe WB, Byrne JC, Leweke FM, Rothermund M, et al. Identification of N-glycosylation changes in the CSF and serum in patients with schizophrenia. J Proteome Res. 2010;9:4476–89.
Kippe JM, Mueller TM, Haroutunian V, Meador-Woodruff JH. Abnormal N-acetylglucosaminyltransferase expression in prefrontal cortex in schizophrenia. Schizophr Res. 2015;166:219–24.
Narayan S, Head SR, Gilmartin TJ, Dean B, Thomas EA. Evidence for disruption of sphingolipid metabolism in schizophrenia. J Neurosci Res. 2009;87:278–88.
Niu L, Matsui M, Zhou SY, Hagino H, Takahashi T, Yoneyama E, et al. Volume reduction of the amygdala in patients with schizophrenia: a magnetic resonance imaging study. Psychiatry Res. 2004;132:41–51.
Aleman A, Kahn RS. Strange feelings: do amygdala abnormalities dysregulate the emotional brain in schizophrenia? Prog Neurobiol. 2005;77:283–98.
Baiano M, Perlini C, Rambaldelli G, Cerini R, Dusi N, Bellani M, et al. Decreased entorhinal cortex volumes in schizophrenia. Schizophr Res. 2008;102:171–80.
Harrison PJ. The neuropathology of schizophrenia. A critical review of the data and their interpretation. Brain. 1999;122:593–624.
Nakagawa F, Schulte BA, Spicer SS. Selective cytochemical demonstration of glycoconjugate-containing terminal N-acetylgalactosamine on some brain neurons. J Comp Neurol. 1986;243:280–90.
Kawakami K, Matsumoto K. Behavioral alterations in mice lacking the gene for tenascin-X. Biol Pharm Bull. 2011;34:590–3.
Liu LL, Wei J, Zhang X, Li XY, Shen Y, Liu SZ, et al. Lack of a genetic association between the TNXB locus and schizophrenia in a Chinese population. Neurosci Lett. 2004;355:149–51.
Tochigi M, Zhang X, Ohashi J, Hibino H, Otowa T, Rogers M, et al. Association study between the TNXB locus and schizophrenia in a Japanese population. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:305–9.
Wang J, Sun S, Zhang L, Wang Z, Ye L, Liu L, et al. Further study of genetic association between the TNXB locus and schizophrenia. Psychiatr Genet. 2011. doi:10.1097/YPG.0b013e3283413398.
Nakata K, Ujike H, Sakai A, Takaki M, Imamura T, Tanaka Y, et al. Association study between the fibronectin gene and schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2003;116B:41–4.
Supriyanto I, Watanabe Y, Mouri K, Shiroiwa K, Ratta-Apha W, Yoshida M, et al. A missense mutation in the ITGA8 gene, a cell adhesion molecule gene, is associated with schizophrenia in Japanese female patients. Progr Neuropsychopharmacol Biol Psychiatry. 2013;40:347–52.
Wang KS, Liu X, Arana TB, Thompson N, Weisman H, Devargas C, et al. Genetic association analysis of ITGB3 polymorphisms with age at onset of schizophrenia. J Mol Neurosci. 2013;51:446–53.
Dow DJ, Huxley-Jones J, Hall JM, Francks C, Maycox PR, Kew JN, et al. ADAMTSL3 as a candidate gene for schizophrenia: gene sequencing and ultra-high density association analysis by imputation. Schizophr Res. 2011;127:28–34.
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7.
Chopra K, Baveja A, Kuhad A. MMPs: a novel drug target for schizophrenia. Exp Opin Ther Targets. 2015;19:77–85.
Wei J, Richbourgh B, Jia T, Liu C. ADAMTS-12: a multifaced metalloproteinase in arthritis and inflammation. Mediators Inflamm. 2014. doi:10.1155/2014/649718.
Farkas N, Lendeckel U, Dobrowolny H, Funke S, Steiner J, Keilhoff G, et al. Reduced density of ADAM 12-immunoreactive oligodendrocytes in the anterior cingulate white matter of patients with schizophrenia. World J Biol Psychiatry. 2010;11:556–66.
Mühleisen TW, Mattheisen M, Strohmaier J, Degenhardt F, Priebe L, Schultz CC, et al. Association between schizophrenia and common variation in neurocan (NCAN), a genetic risk factor for bipolar disorder. Schizophr Res. 2012;138:69–73.
Schultz CC, Mühleisen TW, Nenadic I, Koch K, Wagner G, Schachtzabel C, et al. Common variation in NCAN, a risk factor for bipolar disorder and schizophrenia, influences local cortical folding in schizophrenia. Psychol Med. 2014;44:811–20.
So HC, Fong PY, Chen RY, Hui TC, Ng MY, Cherny SS, et al. Identification of neuroglycan C and interacting partners as potential susceptibility genes for schizophrenia in a southern Chinese population. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:103–13.
Davalieva K, Maleva Kostovska I, Dwork AJ. Proteomics research in schizophrenia. Front Cell Neurosci. 2016. doi:10.3389/fncel.2016.00018.
Schwarz E, Izmailov R, Spain M, Barnes A, Mapes JP, Guest PC, et al. Validation of a blood-based laboratory test to aid in the confirmation of a diagnosis of schizophrenia. Biomark Insights. 2010;5:39–47.
Schwarz E, Guest PC, Rahmoune H, Harris LW, Wang L, Leweke FM, et al. Identification of a biological signature for schizophrenia in serum. Mol Psychiatry. 2012;17:494–502.
Schwarz E, van Beveren NJ, Ramsey J, Leweke FM, Rothermundt M, Bogerts B, et al. Identification of subgroups of schizophrenia patients with changes in either immune or growth factor and hormonal pathways. Schizophr Bull. 2014;40:787–95.
Selkoe DJ. Alzheimer's disease: genotypes, phenotypes, and treatments. Science. 1997;275:630–1.
Schworer R, Zubkova OV, Turnbull JE, Tyler PC. Synthesis of a targeted library of heparan sulfate hexa- to dodecasaccharides as inhibitors of beta-secretase: potential therapeutics for Alzheimer's disease. Chemistry. 2013;19:6817–23.
Patey SJ, Edwards EA, Yates EA, Turnbull JE. Engineered heparins: novel beta-secretase inhibitors as potential Alzheimer's disease therapeutics. Neurodegener Dis. 2008;5:197–9.
DeWitt DA, Silver J, Canning DR, Perry G. Chondroitin sulfate proteoglycans are associated with the lesions of Alzheimer's disease. Exp Neurol. 1993;121:149–52.
Pangalos MN, Shioi J, Efthimiopoulos S, Wu A, Robakis NK. Characterization of appican, the chondroitin sulfate proteoglycan form of the Alzheimer amyloid precursor protein. Neurodegeneration. 1996;5:445–51.
Cui H, Freeman C, Jacobson GA, Small DH. Proteoglycans in the central nervous system: role in development, neural repair, and Alzheimer's disease. IUBMB Life. 2013;65:108–20.
Scholefield Z, Yates EA, Wayne G, Amour A, McDowell W, Turnbull JE. Heparan sulfate regulates amyloid precursor protein processing by BACE1, the Alzheimer's beta-secretase. J Cell Biol. 2003;163:97–107.
Castillo GM, Ngo C, Cummings J, Wight TN, Snow AD. Perlecan binds to the beta-amyloid proteins (Aβ) of Alzheimer's disease, accelerates Aβ fibril formation, and maintains Aβ fibril stability. J Neurochem. 1997;69:2452–65.
Castillo GM, Lukito W, Wight TN, Snow AD. The sulfate moieties of glycosaminoglycans are critical for the enhancement of beta-amyloid protein fibril formation. J Neurochem. 1999;72:1681–7.
Bruinsma IB, te Riet L, Gevers T, ten Dam GB, van Kuppevelt TH, David G, et al. Sulfation of heparan sulfate associated with amyloid-beta plaques in patients with Alzheimer's disease. Acta Neuropathol. 2010;119:211–20.
Laikko I, Larmas M. Adenosine triphosphate in normal and carious human dentine. Arch Oral Biol. 1979;24:15–20.
Watanabe N, Araki W, Chui DH, Makifuchi T, Ihara Y, Tabira T. Glypican-1 as an Abeta binding HSPG in the human brain: its localization in DIG domains and possible roles in the pathogenesis of Alzheimer's disease. FASEB J. 2004;18:1013–5.
Leonova EI, Galzitskaia OV. Role of syndecan-2 in amyloid plaque formation. Mol Biol (Mosk). 2015;49:89–98.
Verbeek MM, Otte-Holler I, van den Born J, van den Heuvel LP, David G, Wesseling P, et al. Agrin is a major heparan sulfate proteoglycan accumulating in Alzheimer's disease brain. Am J Pathol. 1999;155:2115–25.
Snow AD, Sekiguchi R, Nochlin D, Fraser P, Kimata K, Mizutani A, et al. An important role of heparan sulfate proteoglycan (perlecan) in a model system for the deposition and persistence of fibrillar Aβ-amyloid in rat brain. Neuron. 1994;12:219–34.
Snow AD, Kinsella MG, Parks E, Sekiguchi RT, Miller JD, Kimata K, et al. Differential binding of vascular cell-derived proteoglycans (perlecan, biglycan, decorin, and versican) to the beta-amyloid protein of Alzheimer's disease. Arch Biochem Biophys. 1995;320:84–95.
Snow AD, Mar H, Nochlin D, Sekiguchi RT, Kimata K, Koike Y, et al. Early accumulation of heparan sulfate in neurons and in the beta-amyloid protein-containing lesions of Alzheimer's disease and Down's syndrome. Am J Pathol. 1990;137:1253–70.
Snow AD, Sekiguchi RT, Nochlin D, Kalaria RN, Kimata K. Heparan sulfate proteoglycan in diffuse plaques of hippocampus but not of cerebellum in Alzheimer's disease brain. Am J Pathol. 1994;144:337–47.
Snow AD, Mar H, Nochlin D, Kresse H, Wight TN. Peripheral distribution of dermatan sulfate proteoglycans (decorin) in amyloid-containing plaques and their presence in neurofibrillary tangles of Alzheimer's disease. J Histochem Cytochem. 1992;40:105–13.
Pangalos MN, Shioi J, Robakis NK. Expression of the chondroitin sulfate proteoglycans of amyloid precursor (appican) and amyloid precursor-like protein 2. J Neurochem. 1995;65:762–9.
Bruckner G, Hausen D, Hartig W, Drlicek M, Arendt T, Brauer K. Cortical areas abundant in extracellular matrix chondroitin sulphate proteoglycans are less affected by cytoskeletal changes in Alzheimer's disease. Neuroscience. 1999;92:791–805.
Baig S, Wilcock GK, Love S. Loss of perineuronal net N-acetylgalactosamine in Alzheimer's disease. Acta Neuropathol. 2005;110:393–401.
Morawski M, Pavlica S, Seeger G, Grosche J, Kouznetsova E, Schliebs R, et al. Perineuronal nets are largely unaffected in Alzheimer model Tg2576 mice. Neurobiol Aging. 2010;31:1254–6.
Kobayashi K, Emson PC, Mountjoy CQ. Vicia villosa lectin-positive neurones in human cerebral cortex. Loss in Alzheimer-type dementia. Brain Res. 1989;498:170–4.
Suttkus A, Rohn S, Weigel S, Glockner P, Arendt T, Morawski M. Aggrecan, link protein and tenascin-R are essential components of the perineuronal net to protect neurons against iron-induced oxidative stress. Cell Death Dis. 2014. doi:10.1038/cddis.2014.25.
Morawski M, Bruckner G, Jager C, Seeger G, Arendt T. Neurons associated with aggrecan-based perineuronal nets are protected against tau pathology in subcortical regions in Alzheimer's disease. Neuroscience. 2010;169:1347–63.
Yasuhara O, Akiyama H, McGeer EG, McGeer PL. Immunohistochemical localization of hyaluronic acid in rat and human brain. Brain Res. 1994;635:269–82.
Saftig P, Reiss K. The “a disintegrin and metalloproteases” ADAM10 and ADAM17: novel drug targets with therapeutic potential? Eur J Cell Biol. 2011;90(6-7):527–35.
Gatta LB, Albertini A, Ravid R, Finazzi D. Levels of beta-secretase BACE and alpha-secretase ADAM10 mRNAs in Alzheimer hippocampus. Neuroreport. 2002;13:2031–3.
Kim M, Suh J, Romano D, Truong MH, Mullin K, Hooli B, et al. Potential late-onset Alzheimer's disease-associated mutations in the ADAM10 gene attenuate α-secretase activity. Hum Mol Genet. 2009;18:3987–96.
Botella-Lopez A, Burgaya F, Gavin R, Garcia-Ayllon MS, Gomez-Tortosa E, Pena-Casanova J, et al. Reelin expression and glycosylation patterns are altered in Alzheimer's disease. Proc Natl Acad Sci U S A. 2006;103:5573–8.
Herring A, Donath A, Steiner KM, Widera MP, Hamzehian S, Kanakis D, et al. Reelin depletion is an early phenomenon of Alzheimer's pathology. J Alzheimers Dis. 2012;30:963–79.
Chin J, Massaro CM, Palop JJ, Thwin MT, Yu GQ, Bien-Ly N, et al. Reelin depletion in the entorhinal cortex of human amyloid precursor protein transgenic mice and humans with Alzheimer's disease. J Neurosci. 2007;27:2727–33.
Dulabon L, Olson EC, Taglienti MG, Eisenhuth S, McGrath B, Walsh CA, et al. Reelin binds α3β1 integrin and inhibits neuronal migration. Neuron. 2000;27:33–44.
Deane R, Sagare A, Hamm K, Parisi M, Lane S, Finn MB, et al. apoE isoform-specific disruption of amyloid beta peptide clearance from mouse brain. J Clin Invest. 2008;118:4002–13.
Brich J, Shie FS, Howell BW, Li R, Tus K, Wakeland EK, et al. Genetic modulation of tau phosphorylation in the mouse. J Neurosci. 2003;23:187–92.
Wang Q, Klyubin I, Wright S, Griswold-Prenner I, Rowan MJ, Anwyl R. Alpha v integrins mediate beta-amyloid induced inhibition of long-term potentiation. Neurobiol Aging. 2008;29:1485–93.
Wright S, Malinin NL, Powell KA, Yednock T, Rydel RE, Griswold-Prenner I. α2β1 and αVβ1 integrin signaling pathways mediate amyloid-beta-induced neurotoxicity. Neurobiol Aging. 2007;28:226–37.
Zenaro E, Pietronigro E, Della Bianca V, Piacentino G, Marongiu L, Budui S, et al. Neutrophils promote Alzheimer's disease-like pathology and cognitive decline via LFA-1 integrin. Nat Med. 2015;21:880–6.
Young-Pearse TL, Chen AC, Chang R, Marquez C, Selkoe DJ. Secreted APP regulates the function of full-length APP in neurite outgrowth through interaction with integrin beta1. Neural Dev. 2008. doi:10.1186/1749-8104-3-15.
Vegh MJ, Heldring CM, Kamphuis W, Hijazi S, Timmerman AJ, Li KW, et al. Reducing hippocampal extracellular matrix reverses early memory deficits in a mouse model of Alzheimer's disease. Acta Neuropathol Commun. 2014. doi:10.1186/s40478-014-0076-z.
Hondius DC, van Nierop P, Li KW, Hoozemans JJ, van der Schors RC, van Haastert ES, et al. Profiling the human hippocampal proteome at all pathologic stages of Alzheimer's disease. Alzheimers Dement. 2016;12:654–68.
Donovan LE, Higginbotham L, Dammer EB, Gearing M, Rees HD, Xia Q, et al. Analysis of a membrane-enriched proteome from postmortem human brain tissue in Alzheimer's disease. Proteomics Clin Appl. 2012;6:201–11.
Andreev VP, Petyuk VA, Brewer HM, Karpievitch YV, Xie F, Clarke J, et al. Label-free quantitative LC-MS proteomics of Alzheimer's disease and normally aged human brains. J Proteome Res. 2012;11:3053–67.
Musunuri S, Wetterhall M, Ingelsson M, Lannfelt L, Artemenko K, Bergquist J, et al. Quantification of the brain proteome in Alzheimer's disease using multiplexed mass spectrometry. J Proteome Res. 2014;13:2056–68.
Ho Kim J, Franck J, Kang T, Heinsen H, Ravid R, Ferrer I, et al. Proteome-wide characterization of signalling interactions in the hippocampal CA4/DG subfield of patients with Alzheimer's disease. Sci Rep. 2015. doi:10.1038/srep11138.
Minjarez B, Calderon-Gonzalez KG, Rustarazo ML, Herrera-Aguirre ME, Labra-Barrios ML, Rincon-Limas DE, et al. Identification of proteins that are differentially expressed in brains with Alzheimer's disease using iTRAQ labeling and tandem mass spectrometry. J Proteomics. 2016;139:103–21.
Sultana R, Boyd-Kimball D, Cai J, Pierce WM, Klein JB, Merchant M, et al. Proteomics analysis of the Alzheimer's disease hippocampal proteome. J Alzheimers Dis. 2007;11:153–64.
Gozal YM, Duong DM, Gearing M, Cheng D, Hanfelt JJ, Funderburk C, et al. Proteomics analysis reveals novel components in the detergent-insoluble subproteome in Alzheimer's disease. J Proteome Res. 2009;8:5069–79.
Shah DJ, Rohlfing F, Anand S, Johnson WE, Alvarez MT, Cobell J, et al. Discovery and subsequent confirmation of novel serum biomarkers diagnosing Alzheimer's sisease. J Alzheimers Dis. 2015;49(2):317–27.
Perneczky R, Guo LH. Plasma proteomics biomarkers in Alzheimer's disease: latest advances and challenges. Methods Mol Biol. 2016;1303:521–9.
Brinkmalm A, Portelius E, Ohrfelt A, Brinkmalm G, Andreasson U, Gobom J, et al. Explorative and targeted neuroproteomics in Alzheimer's disease. Biochim Biophys Acta. 1854;2015:769–78.
Schedin-Weiss S, Winblad B, Tjernberg LO. The role of protein glycosylation in Alzheimer disease. FEBS J. 2014;281:46–62.
Kizuka Y, Kitazume S, Fujinawa R, Saito T, Iwata N, Saido TC, et al. An aberrant sugar modification of BACE1 blocks its lysosomal targeting in Alzheimer's disease. EMBO Mol Med. 2015;7:175–89.
Palmigiano A, Barone R, Sturiale L, Sanfilippo C, Bua RO, Romeo DA, et al. CSF N-glycoproteomics for early diagnosis in Alzheimer's disease. J Proteomics. 2016;131:29–37.
Maguire TM, Gillian AM, O'Mahony D, Coughlan CM, Dennihan A, Breen KC. A decrease in serum sialyltransferase levels in Alzheimer's disease. Neurobiol Aging. 1994;15:99–102.
Maguire TM, Breen KC. A decrease in neural sialyltransferase activity in Alzheimer's disease. Dementia. 1995;6:185–90.
Fodero LR, Saez-Valero J, Barquero MS, Marcos A, McLean CA, Small DH. Wheat germ agglutinin-binding glycoproteins are decreased in Alzheimer's disease cerebrospinal fluid. J Neurochem. 2001;79:1022–6.
Taniguchi M, Okayama Y, Hashimoto Y, Kitaura M, Jimbo D, Wakutani Y, et al. Sugar chains of cerebrospinal fluid transferrin as a new biological marker of Alzheimer's disease. Dement Geriatr Cogn Disord. 2008;26:117–22.
Chen CC, Engelborghs S, Dewaele S, Le Bastard N, Martin JJ, Vanhooren V, et al. Altered serum glycomics in Alzheimer disease: a potential blood biomarker? Rejuvenation Res. 2010;13:439–44.
Korolainen MA, Nyman TA, Aittokallio T, Pirttila T. An update on clinical proteomics in Alzheimer's research. J Neurochem. 2010;112:1386–414.
Papassotiropoulos A, Fountoulakis M, Dunckley T, Stephan DA, Reiman EM. Genetics, transcriptomics, and proteomics of Alzheimer's disease. J Clin Psychiatry. 2006;67:652–70.
Di Domenico F, Sultana R, Barone E, Perluigi M, Cini C, Mancuso C, et al. Quantitative proteomics analysis of phosphorylated proteins in the hippocampus of Alzheimer's disease subjects. J Proteomics. 2011;74:1091–103.
Zhang J, Goodlett DR, Peskind ER, Quinn JF, Zhou Y, Wang Q, et al. Quantitative proteomic analysis of age-related changes in human cerebrospinal fluid. Neurobiol Aging. 2005;26:207–27.
Muller T, Jung K, Ullrich A, Schrotter A, Meyer HE, Stephan C, et al. Disease state, age, sex, and post-mortem time-dependent expression of proteins in AD vs. control frontal cortex brain samples. Curr Alzheimer Res. 2008;5:562–71.
Riederer IM, Schiffrin M, Kovari E, Bouras C, Riederer BM. Ubiquitination and cysteine nitrosylation during aging and Alzheimer's disease. Brain Res Bull. 2009;80:233–41.
Newman SF, Sultana R, Perluigi M, Coccia R, Cai J, Pierce WM, et al. An increase in S-glutathionylated proteins in the Alzheimer's disease inferior parietal lobule, a proteomics approach. J Neurosci Res. 2007;85:1506–14.
Hynd MR, Lewohl JM, Scott HL, Dodd PR. Biochemical and molecular studies using human autopsy brain tissue. J Neurochem. 2003;85:543–62.
McCullumsmith RE, Hammond JH, Shan D, Meador-Woodruff JH. Postmortem brain: an underutilized substrate for studying severe mental illness. Neuropsychopharmacology. 2014;39:65–87.
Piehowski PD, Petyuk VA, Orton DJ, Xie F, Moore RJ, Ramirez-Restrepo M, et al. Sources of technical variability in quantitative LC-MS proteomics: human brain tissue sample analysis. J Proteome Res. 2013;12:2128–37.
Chiou S-H, Wu S-H. Evaluation of commonly used electrophoretic methods for the analysis of proteins and peptides and their application to biotechnology. Anal Chim Acta. 1999;383:47–60.
Rabilloud T, Chevallet M, Luche S, Lelong C. Two-dimensional gel electrophoresis in proteomics: past, present and future. J Proteomics. 2010;73:2064–77.
Cellulaire B. Two-dimensional gel electrophoresis in proteomics: old, old fashioned, but it still climbs up the mountains. Proteomics. 2002;2:3–10.
Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotechnol. 2003;21:255–61.
Acknowledgments
This work was supported by NIH grants P41GM104602, R21CA177479, and R01MH105608.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Published in the topical collection Glycomics Glycoproteomics and Allied Topics with guest editors Yehia Mechref and David Muddiman.
Rights and permissions
About this article
Cite this article
Sethi, M.K., Zaia, J. Extracellular matrix proteomics in schizophrenia and Alzheimer’s disease. Anal Bioanal Chem 409, 379–394 (2017). https://doi.org/10.1007/s00216-016-9900-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-016-9900-6