Abstract
Reversible protein acetylation catalyzed by histone acetyltransferases (HATs) and histone deacetylases (HDACs) is an essential post-translational modification (PTM) mechanism which correlates largely with epigenetic gene regulation such as transcriptional activation, DNA replication, histone deposition, and DNA repair. Dysfunction of histone acetylation and the aberrant activity of HATs/HDACs is often associated with the pathogenesis of numerous diseases, especially cancer. Therefore, developing potent and specific analytical methods for HATs/HDACs is important for fundamental biochemical research, disease diagnosis and treatment, and drug development. This paper briefly summarizes the general design strategies used in HAT/HDAC sensors, gives a systematic overview of recent advances in the analytical methods for HAT/HDAC enzymatic analysis, classifies these methods by their biorecognition mechanisms and relative applications either in vitro or in living cells, then outlines challenges faced by these bioanalytical methods and offers perspectives on future developments.

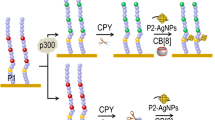
Reversible acetylation modification process and the general sensing mechanisms of protein acetylation-related enzymes (PAREs) activity





Similar content being viewed by others
References
Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–80.
Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327(5968):1000–4.
Biel M, Wascholowski V, Giannis A. Epigenetics-an epicenter of gene regulation: histones and histone-modifying enzymes. Angew Chem Int Ed. 2005;44(21):3186–216.
Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–40.
Dekker FJ, Haisma HJ. Histone acetyl transferases as emerging drug targets. Drug Discov Today. 2009;14(19):942–8.
Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov. 2014;13(9):673–91.
Gryder BE, Sodji QH, Oyelere AK. Targeted cancer therapy: giving histone deacetylase inhibitors all they need to succeed. Future Med Chem. 2012;4(4):505–24.
Ratner M. Small biotech steers HDAC inhibitor to clinic. Nat Biotechnol. 2014;32(9):853–4.
Yang XJ, Seto EY. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26(37):5310–8.
Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11(5):384–400.
Kuninger D, Lundblad J, Semirale A, Rotwein P. A non-isotopic in vitro assay for histone acetylation. J Biotechnol. 2007;131(3):253–60.
Ghadiali JE, Lowe SB, Stevens MM. Quantum-dot-based FRET detection of histone acetyltransferase activity. Angew Chem Int Ed. 2011;123(15):3479–82.
Zhen Z, Tang L, Long H, Jiang J. Enzymatic immuno-assembly of gold nanoparticles for visualized activity screening of histone-modifying enzymes. Anal Chem. 2012;84(8):3614–20.
Wu H, Liu S, Zhu W, Jiang J, Shen G, Yu R. A sensitive electrochemical biosensor for detection of histone deacetylase activity using an acetylated peptide. Electroanalysis. 2012;24(12):2365–70.
Zhang S, Wu H, Huan S, Zhang X, Shen G, Yu R. Gold nanoparticle based fluorescence resonance energy transfer immunoassay for the detection of the histone deacetylase activity using a fluorescent peptide probe. Anal Lett. 2013;46(13):2029–39.
Wegener D, Hildmann C, Riester D, Schwienhorst A. Improved fluorogenic histone deacetylase assay for high-throughput-screening applications. Anal Biochem. 2003;321(2):202–8.
Wegener D, Wirsching F, Riester D, Schwienhorst A. A fluorogenic histone deacetylase assay well suited for high-throughput activity screening. Chem Biol. 2003;10(1):61–8.
Marcotte PA, Richardson PL, Guo J, Barrett LW, Xu N, Gunasekera A, et al. Fluorescence assay of SIRT protein deacetylases using an acetylated peptide substrate and a secondary trypsin reaction. Anal Biochem. 2004;332(1):90–9.
Dose A, Jost JO, Spiess AC, Henklein P, Beyermann M, Schwarzer D. Facile synthesis of colorimetric histone deacetylase substrates. Chem Commun. 2012;48(76):9525–7.
Ueki N, Lee S, Sampson NS, Hayman MJ. Selective cancer targeting with prodrugs activated by histone deacetylases and a tumour-associated protease. Nat Commun. 2013;4(4):2735.
Wang Y, Chen Y, Wang H, Cheng Y, Zhao X. Specific turn-on fluorescent probe with aggregation-induced emission characteristics for SIRT1 modulator screening and living-cell imaging. Anal Chem. 2015;87(10):5046–9.
Han Y, Li P, Xu Y, Li H, Song Z, Nie Z, et al. Fluorescent nanosensor for probing histone acetyltransferase activity based on acetylation protection and magnetic graphitic nanocapsules. Small. 2015;11(7):877–85.
Dhara K, Hori Y, Baba R, Kikuchi K. A fluorescent probe for detection of histone deacetylase activity based on aggregation-induced emission. Chem Commun. 2012;48(94):11534–6.
Minoshima M, Matsumoto T, Kikuchi K. Development of a fluorogenic probe based on a DNA staining dye for continuous monitoring of the histone deacetylase reaction. Anal Chem. 2014;86(15):7925–30.
Han Y, Li H, Hu Y, Li P, Wang H, Nie Z, et al. Time-resolved luminescence biosensor for continuous activity detection of protein acetylation-related enzymes based on DNA-sensitized terbium(III) probes. Anal Chem. 2015;87(18):9179–85.
Heltweg B, Dequiedt F, Verdin E, Jung M. Nonisotopic substrate for assaying both human zinc and NAD+-dependent histone deacetylases. Anal Biochem. 2003;319(1):42–8.
Heltweg B, Jung M. A homogeneous nonisotopic histone deacetylase activity assay. J Biomol Screen. 2003;8(1):89–95.
Baba R, Hori Y, Kikuchi K. Intramolecular long-distance nucleophilic reactions as a rapid fluorogenic switch applicable to the detection of enzymatic activity. Chem Eur J. 2015;21(12):4695–702.
Baba R, Hori Y, Mizukami S, Kikuchi K. Development of a fluorogenic probe with a transesterification switch for detection of histone deacetylase activity. J Am Chem Soc. 2012;134(35):14310–3.
Rooker DR, Buccella D. Real-time detection of histone deacetylase activity with a small molecule fluorescent and spectrophotometric probe. Chem Sci. 2015. doi:10.1039/c5sc02704g.
Wu J, Zheng YG. Fluorescent reporters of the histone acetyltransferase. Anal Biochem. 2008;380(1):106–10.
Wen Q, Gu Y, Tang L, Yu R, Jiang J. Peptide-templated gold nanocluster beacon as a sensitive, label-free sensor for protein post-translational modification enzymes. Anal Chem. 2013;85(24):11681–5.
Zheng D, Ye Z, Yang S, Li Y, He X, Li G. An electrochemical method to evaluate p53 C-terminal domain acetylation on its DNA binding ability. Electrochem Commun. 2014;49:30–3.
Minaker SA, Daze KD, Ma MC, Hof F. Antibody-free reading of the histone code using a simple chemical sensor array. J Am Chem Soc. 2012;134(28):11674–80.
Kim Y, Tanner KG, Denu JM. A continuous, nonradioactive assay for histone acetyltransferases. Anal Biochem. 2000;280(2):308–14.
Trievel RC, Li FY, Marmorstein R. Application of a fluorescent histone acetyltransferase assay to probe the substrate specificity of the human p300/CBP-associated factor. Anal Biochem. 2000;287(2):319–28.
Gao T, Yang C, Zheng YG. Comparative studies of thiol-sensitive fluorogenic probes for HAT assays. Anal Bioanal Chem. 2013;405(4):1361–71.
Chen S, Li Y, Hu Y, Han Y, Huang Y, Nie Z, et al. Nucleic acid-mimicking coordination polymer for label-free fluorescent activity assay of histone acetyltransferases. Chem Commun. 2015;51(21):4469–72.
Hu Y, Chen S, Han Y, Chen H, Wang Q, Nie Z, et al. Unique electrocatalytic activity of nucleic acid-mimicking coordination polymer for sensitive detection of coenzyme A and histone acetyltransferase activity. Chem Commun. 2015. doi:10.1039/c5cc06593c.
Riester D, Hildmann C, Schwienhorst A, Meyer-Almes FJ. Histone deacetylase inhibitor assay based on fluorescence resonance energy transfer. Anal Biochem. 2007;362(1):136–41.
Riester D, Hildmann C, Haus P, Galetovic A, Schober A, Schwienhorst A, et al. Non-isotopic dual parameter competition assay suitable for high-throughput screening of histone deacetylases. Bioorg Med Chem Lett. 2009;19(13):3651–6.
Xie N, Elangwe EN, Asher S, Zheng YG. A dual-mode fluorescence strategy for screening HAT modulators. Bioconjug Chem. 2009;20(2):360–6.
Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399(6735):491–6.
Yang X. Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays. 2004;26(10):1076–87.
Kanno T, Kanno Y, Siegel RM, Jang MK, Lenardo MJ, Ozato K. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol Cell. 2004;13(1):33–43.
Sasaki K, Ito T, Nishino N, Khochbin S, Yoshida M. Real-time imaging of histone H4 hyperacetylation in living cells. Proc Natl Acad Sci U S A. 2009;106(38):16257–62.
Dancy BM, Crump NT, Peterson DJ, Mukherjee C, Bowers EM, Ahn YH, et al. Live-cell studies of p300/CBP histone acetyltransferase activity and inhibition. Chem Biol Chem. 2012;13(14):2113–21.
Ito T, Umehara T, Sasaki K, Nakamura Y, Nishino N, Terada T, et al. Real-time imaging of histone H4K12-specific acetylation determines the modes of action of histone deacetylase and bromodomain inhibitors. Chem Biol. 2011;18(4):495–507.
Nakaoka S, Sasaki K, Ito A, Nakao Y, Yoshida M. A genetically encoded FRET probe to detect intranucleosomal histone H3K9 or H3K14 acetylation using BRD4, a BET family member. ACS Chem Biol. 2015. doi:10.1021/cb501046t.
Hayashi-Takanaka Y, Yamagata K, Wakayama T, Stasevich TJ, Kainuma T, Tsurimoto T, et al. Tracking epigenetic histone modifications in single cells using Fab-based live endogenous modification labeling. Nucleic Acids Res. 2011;39(15):6475–88.
Sato Y, Mukai M, Ueda J, Muraki M, Stasevich TJ, Horikoshi N, et al. Genetically encoded system to track histone modification in vivo. Sci Rep. 2013;3:2436.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (Nos. 21222507, 21175036, 21235002, and 21475038), the National Basic Research Program of China (973 Program, No. 2011CB911002), the Foundation for Innovative Research Groups of NSFC (Grant 21221003), and the Natural Science Foundation of Hunan Province (No. 2015JJ1005).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no potential conflict of interest.
Additional information
Published in the topical collection featuring Young Investigators in Analytical and Bioanalytical Science with guest editors S. Daunert, A. Baeumner, S. Deo, J. Ruiz Encinar, and L. Zhang.
Rights and permissions
About this article
Cite this article
Li, P., Han, Y., Li, Y. et al. Bioanalytical approaches for the detection of protein acetylation-related enzymes. Anal Bioanal Chem 408, 2659–2668 (2016). https://doi.org/10.1007/s00216-016-9304-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-016-9304-7