Abstract
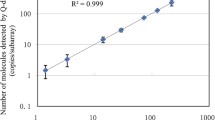
Plasmid calibrators are increasingly applied for polymerase chain reaction (PCR) analysis of genetically modified organisms (GMOs). To evaluate the commutability between plasmid DNA (pDNA) and genomic DNA (gDNA) as calibrators, a plasmid molecule, pBSTopas, was constructed, harboring a Topas 19/2 event-specific sequence and a partial sequence of the rapeseed reference gene CruA. Assays of the pDNA showed similar limits of detection (five copies for Topas 19/2 and CruA) and quantification (40 copies for Topas 19/2 and 20 for CruA) as those for the gDNA. Comparisons of plasmid and genomic standard curves indicated that the slopes, intercepts, and PCR efficiency for pBSTopas were significantly different from CRM Topas 19/2 gDNA for quantitative analysis of GMOs. Three correction methods were used to calibrate the quantitative analysis of control samples using pDNA as calibrators: model a, or coefficient value a (Cva); model b, or coefficient value b (Cvb); and the novel model c or coefficient formula (Cf). Cva and Cvb gave similar estimated values for the control samples, and the quantitative bias of the low concentration sample exceeded the acceptable range within ±25 % in two of the four repeats. Using Cfs to normalize the Ct values of test samples, the estimated values were very close to the reference values (bias −13.27 to 13.05 %). In the validation of control samples, model c was more appropriate than Cva or Cvb. The application of Cf allowed pBSTopas to substitute for Topas 19/2 gDNA as a calibrator to accurately quantify the GMO.

ᅟ



Similar content being viewed by others
References
Block A, Debode F, Grohmann L, Hulin J, Taverniers I, Kluga L, Barbau-Piednoir E, Broeders S, Huber I, Van den Bulcke M, Heinze P, Berben G, Busch U, Roosens N, Janssen E, Zel J, Gruden K, Morisset D (2013) BMC Bioinforma 14:256
Holst-Jensen A, Bertheau Y, De LM, Grohmann L, Hamels S, Hougs L, Morisset D, Pecoraro S, Pla M, Van BM, Wulff D (2012) Biotechnol Adv 30:1318–1335
Ciabatti I, Marchesi U, Froiio A, Patern A, Ruggeri M, Amaddeo D (2005) Vet Res Commun 29:31–34
Lauwaars M, Anklam E (2004) Accred Qual Assur J Qual Comp Reliab Chem Meas 9:253–258
Burns M, Valdivia H (2007) Eur Food Res Technol 226:7–18
Holst-Jensen A (2009) Biotechnol Adv 27:1071–1082
Regulation EC No 787/2004 (2004) Off J Eur Communities L348:18–21
Regulation EC No 619/2011 (2011) Off J Eur Communities L166:9–13
Gancberg D, Corbisier P, De Andrade Silva E, Mazoua S, Merveillie A, Tumba M, Trapmann S (2009) Certification of reference materials of maize seed powder with different mass fractions of genetically modified 98140 maize ERM®-BF427
James C (2014) ISAAA Brief No. 46
Taverniers I, Van BE, De LM (2001) Eur Food Res Technol 66:469–472
Kuribara H, Shindo Y, Matsuoka T, Takubo K, Futo S, Aoki N, Hirao T, Akiyama H, Goda Y, Toyoda M, Hino A (2002) J AOAC Int 85:1077–1089
Shindo Y, Kuribara H, Matsuoka T, Futo S, Sawada C, Shono J, Akiyama H, Goda Y, Toyoda M, Hino A (2002) J AOAC Int 85:1119–1126
Pardigol A, Guillet S, Popping B (2003) Eur Food Res Technol 216:412–420
Taverniers I, Van BE, De LM (2004) Anal Bioanal Chem 378:1198–1207
Lievens A, Bellocchi G, De BD, Moens W, Savini C, Mazzara M, Van EG, Van BM (2010) Anal Bioanal Chem 396:2165–2173
Taverniers I, Windels P, Vaitilingom M, Milcamps A, Van BE, Van EG, De LM (2005) J Agric Food Chem 53:3041–3052
Jeynov B, De Andrade E, Broothaerts W, Corbisier P, Mazoua S, Merveillie A, Trapmann S, Emons H (2011) Certification of plasmid DNA containing NK603 maize DNA fragments—certified reference material ERM®-AD415
Caprioara-Buda M, Meyer W, Jeynov B, Corbisier P, Trapmann S, Emons H (2012) Anal Bioanal Chem 404:29–42
Corbisier P, Broeders S, Charels D, Trapmann S, Vincent S, Emons H (2007) Certification of plasmidic DNA containing MON 810 maize DNA fragments, ERM®-AD413
Block A, Schwarz G (2003) Eur Food Res Technol 216:421–427
Burns M, Corbisier P, Wiseman G, Valdivia H, McDonald P, Bowler P, Ohara K, Schimmel H, Charels D, Damant A, Harris N (2006) Eur Food Res Technol 224:249–258
Zhang H, Yang L, Guo J, Li X, Jiang L, Zhang D (2008) J Agric Food Chem 56:5514–5520
Yang L, Guo J, Pan A, Zhang H, Zhang K, Wang Z, Zhang D (2007) J Agric Food Chem 55:15–24
Cao Y, Wu G, Wu Y, Nie S, Zhang L, Lu C (2011) J Agric Food Chem 59:8550–8559
Li X, Wu Y, Zhang L, Cao Y, Li Y, Li J, Zhu L, Wu G (2014) Anal Biochem 451:18–24
Community Reference Laboratory For GM Food And Feed (2011) Event-specific method for the quantification of oilseed rape Topas 19/2 using real-time PCR
Wu G, Wu Y, Xiao L, Lu C (2009) Food Chem 112:232–238
Arumuganathan K, Earle E (1991) Plant Mol Biol Report 9:208–218
ENGL (2011) Verification of analytical methods for GMO testing when implementing inter-laboratory validated methods
Toyota A, Akiyama H, Sugimura M, Watanabe T, Kikuchi H, Kanamori H, Hino A, Esaka M, Maitani T (2006) Biosci Biotechnol Biochem 70:821–827
Wang X, Teng D, Yang Y, Tian F, Guan Q, Wang J (2011) Appl Microbiol Biotechnol 90:721–731
ENGL (2008) Definition of minimum performance requirements for analytical methods of GMO testing. European Network of GMO Laboratories
Acknowledgments
This research was supported by grants from the Public Welfare Industry-Special Research Project of Ministry of Environmental Protection (201109028), the National Major Special Project for the Development of Transgenic Organisms (grant nos. 2014ZX08012-003, 2014ZX08012-002B, and 2014ZX08012-005), the Special Fund for Basic Scientific Research of Central Colleges, South-Central University for Nationalities (grant no. CZQ13009), and the Major Research Project of CAAS Science and Technology Innovation Program.
Author information
Authors and Affiliations
Corresponding author
Additional information
Li Zhang and Yuhua Wu contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 204 kb)
Rights and permissions
About this article
Cite this article
Zhang, L., Wu, Y., Wu, G. et al. Correction of the lack of commutability between plasmid DNA and genomic DNA for quantification of genetically modified organisms using pBSTopas as a model. Anal Bioanal Chem 406, 6385–6397 (2014). https://doi.org/10.1007/s00216-014-8056-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-014-8056-5