Abstract
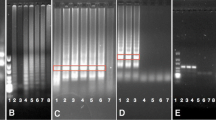
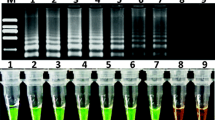
The requirement to monitor the presence of genetically modified organisms (GMO) in a variety of marked products has generated an increasing demand for reliable, rapid, and time and cost-effective analytical methods. Here we report an on-site method for rapid detection of cauliflower mosaic virus promoter (CaMV 35S), a common element present in most GMO, using cross-priming amplification (CPA) technology. Detection was achieved using a DNA-based contamination-proof strip biosensor. The limit of detection was 30 copies for the pBI121 plasmid containing the CaMV 35S gene. The certified reference sample of GM maize line MON810 was detectable even at the low relative mass concentration of 0.05 %. The developed CPA method had high specificity for the CaMV 35S gene, as compared with other GM lines not containing this gene and non-GM products. The method was further validated using nine real-world samples, and the results were confirmed by real-time PCR analysis. Because of its simplicity, rapidity, and high sensitivity, this method of detecting the CaMV 35S gene has great commercial prospects for rapid GMO screening of high-consumption food and agriculture products.





Similar content being viewed by others
References
Hails RS (2000) Genetically modified plants–the debate continues. Trends in Ecology and Evolution 15(1):14–18
Wolfenbarger LL, Phifer PR (2000) The ecological risks and benefits of genetically engineered plants. Science 290(5499):2088–2093
Kaur J, Radu S, Ghazali FM, Kqueen CY (2010) Real-time PCR-based detection and quantification of genetically modified maize in processed feeds commercialised in Malaysia. Food Control 21(11):1536–1544. doi:10.1016/j.foodcont.2010.03.018
Carter CA, Gruère GP (2006) International approval and labeling regulations of genetically modified food in major trading countries. In: Regulating agricultural biotechnology: economics and policy. Springer, pp 459–480
Ahmed FE (2002) Detection of genetically modified organisms in foods. Trends Biotechnol 20(5):215–223
Akiyama H, Nakamura F, Yamada C, Nakamura K, Nakajima O, Kawakami H, Harikai N, Furui S, Kitta K, Teshima R (2009) A screening method for the detection of the 35S promoter and the nopaline synthase terminator in genetically modified organisms in a real-time multiplex polymerase chain reaction using high-resolution melting-curve analysis. Biol Pharm Bull 32(11):1824–1829
Guertler P, Paul V, Albrecht C, Meyer HH (2009) Sensitive and highly specific quantitative real-time PCR and ELISA for recording a potential transfer of novel DNA and Cry1Ab protein from feed into bovine milk. Anal Bioanal Chem 393(6–7):1629–1638
Huang X, Hou L, Xu X, Chen H, Ji H, Zhu S (2011) One-PCR-tube approach for in situ DNA isolation and detection. Analyst 136(20):4254–4259
Gryson N (2010) Effect of food processing on plant DNA degradation and PCR-based GMO analysis: a review. Anal Bioanal Chem 396(6):2003–2022
Lauri A, Mariani PO (2009) Potentials and limitations of molecular diagnostic methods in food safety. Genes and Nutrition 4(1):1–12
Kumar R (2012) Development of Dipsticks for Simultaneous Detection of Vip3A and Cry1Ab/Cry1Ac Transgenic Proteins. J AOAC Int 95(4):1131–1137
Kumar R, Singh CK, Kamle S, Sinha RP, Bhatnagar RK, Kachru DN (2010) Development of nanocolloidal gold based immunochromatographic assay for rapid detection of transgenic vegetative insecticidal protein in genetically modified crops. Food Chem 122(4):1298–1303
Kumar R (2011) A Quantitative Immunopolymerase Chain Reaction Method for Detection of Vegetative Insecticidal Protein in Genetically Modified Crops. J Agric Food Chem 59(19):10448–10453
Trantakis IA, Spaniolas S, Kalaitzis P, Ioannou PC, Tucker GA, Christopoulos TK (2012) Dipstick Test for DNA-Based Food Authentication. Application to Coffee Authenticity Assessment. J Agric Food Chem 60(3):713–717
Tomlinson J, Dickinson M, Boonham N (2010) Rapid detection of Phytophthora ramorum and P. kernoviae by two-minute DNA extraction followed by isothermal amplification and amplicon detection by generic lateral flow device. Phytopathology 100(2):143–149
Walker GT, Fraiser MS, Schram JL, Little MC, Nadeau JG, Malinowski DP (1992) Strand displacement amplification—an isothermal, in vitro DNA amplification technique. Nucleic Acids Res 20(7):1691–1696
Malek RSLT (1995) NASBA. Nat Biotechnol 13:563–564. doi:10.1038/nbt0695-563
Fire A, Xu S-Q (1995) Rolling replication of short DNA circles. Proc Natl Acad Sci 92(10):4641–4645
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28(12):e63–e63
Vincent M, Xu Y, Kong H (2004) Helicase-dependent isothermal DNA amplification. EMBO Rep 5(8):795–800
Xu G, Hu L, Zhong H, Wang H, Yusa S-i, Weiss TC, Romaniuk PJ, Pickerill S, You Q (2012) Cross priming amplification: mechanism and optimization for isothermal DNA amplification. Scientific reports 2
Fang R, Li X, Hu L, You Q, Li J, Wu J, Xu P, Zhong H, Luo Y, Mei J (2009) Cross-priming amplification for rapid detection of Mycobacterium tuberculosis in sputum specimens. J Clin Microbiol 47(3):845–847
Yulong Z, Xia Z, Hongwei Z, Wei L, Wenjie Z, Xitai H (2010) Rapid and sensitive detection of Enterobacter sakazakii by cross-priming amplification combined with immuno-blotting analysis. Mol Cell Probes 24(6):396–400. doi:10.1016/j.mcp.2010.09.001
Zhang J, Tian Q, Zhu SF, Zhao WJ, Liu FQ (2012) Rapid on-site detection of Acidovorax citrulli by cross-priming amplification. Mol Cell Probes 26(4):175–176. doi:10.1016/j.mcp.2012.03.010
Chow WHA, McCloskey C, Tong Y, Hu L, You Q, Kelly CP, Kong H, Tang Y-W, Tang W (2008) Application of Isothermal Helicase-Dependent Amplification with a Disposable Detection Device in a Simple Sensitive Stool Test for Toxigenic Clostridium difficile. J Mol Diagn 10(5):452–458
Guan X, Guo J, Shen P, Yang L, Zhang D (2010) Visual and rapid detection of two genetically modified soybean events using loop-mediated isothermal amplification method. Food Anal Methods 3(4):313–320
Lee D, La Mura M, Allnutt T, Powell W (2009) Detection of genetically modified organisms (GMOs) using isothermal amplification of target DNA sequences. BMC Biotechnol 9(1):7
Iwamoto T, Sonobe T, Hayashi K (2003) Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J Clin Microbiol 41(6):2616–2622
Mori Y, Nagamine K, Tomita N, Notomi T (2001) Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem Biophys Res Commun 289(1):150–154
Mori N, Motegi Y, Shimamura Y, Ezaki T, Natsumeda T, Yonekawa T, Ota Y, Notomi T, Nakayama T (2006) Development of a new method for diagnosis of rubella virus infection by reverse transcription-loop-mediated isothermal amplification. J Clin Microbiol 44(9):3268–3273
Borst A, Box A, Fluit A (2004) False-positive results and contamination in nucleic acid amplification assays: suggestions for a prevent and destroy strategy. Eur J Clin Microbiol Infect Dis 23(4):289–299
Mano J, Yanaka Y, Ikezu Y, Onishi M, Futo S, Minegishi Y, Ninomiya K, Yotsuyanagi Y, Spiegelhalter F, Akiyama H (2011) Practicable group testing method to evaluate weight/weight GMO content in maize grains. J Agric Food Chem 59(13):6856–6863
Mano J, Shigemitsu N, Futo S, Akiyama H, Teshima R, Hino A, Furui S, Kitta K (2008) Real-time PCR array as a universal platform for the detection of genetically modified crops and its application in identifying unapproved genetically modified crops in Japan. J Agric Food Chem 57(1):26–37
Grohmann L, Brünen-Nieweler C, Nemeth A, Waiblinger H-U (2009) Collaborative trial validation studies of real-time PCR-based GMO screening methods for detection of the bar gene and the ctp2-cp4epsps construct. J Agric Food Chem 57(19):8913–8920
Kodama T, Kuribara H, Minegishi Y, Futo S, Watai M, Sawada C, Watanabe T, Akiyama H, Maitani T, Teshima R (2008) Evaluation of modified PCR quantitation of genetically modified maize and soybean using reference molecules: interlaboratory study. J AOAC Int 92(1):223–233
Huang H-Y, Pan T-M (2004) Detection of genetically modified maize MON810 and NK603 by multiplex and real-time polymerase chain reaction methods. J Agric Food Chem 52(11):3264–3268
Nascimento V, Von Pinho E, Von Pinho R, Do Nascimento Júnior A (2012) Detection limits of the strip test and PCR for genetically modified corn in Brazil. Genet Mol Res 11(3):2497–2505
Van den Bulcke M, De Schrijver A, De Bernardi D, Devos Y, MbongoMbella G, Casi AL, Moens W, Sneyers M (2007) Detection of genetically modified plant products by protein strip testing: an evaluation of real-life samples. Eur Food Res Technol 225(1):49–57
Acknowledgments
We are grateful for the support of the Important National Science and Technology Specific Projects (2013ZX08012-001) and AQSIQ Non-profit Industry Research Projects (20141004).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 1.32 mb)
Rights and permissions
About this article
Cite this article
Huang, X., Zhai, C., You, Q. et al. Potential of cross-priming amplification and DNA-based lateral-flow strip biosensor for rapid on-site GMO screening. Anal Bioanal Chem 406, 4243–4249 (2014). https://doi.org/10.1007/s00216-014-7791-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-014-7791-y