Abstract
Systems biology requires knowledge of the absolute amounts of proteins in order to model biological processes and simulate the effects of changes in specific model parameters. Quantification concatamers (QconCATs) are established as a method to provide multiplexed absolute peptide standards for a set of target proteins in isotope dilution standard experiments. Two or more quantotypic peptides representing each of the target proteins are concatenated into a designer gene that is metabolically labelled with stable isotopes in Escherichia coli or other cellular or cell-free systems. Co-digestion of a known amount of QconCAT with the target proteins generates a set of labelled reference peptide standards for the unlabelled analyte counterparts, and by using an appropriate mass spectrometry platform, comparison of the intensities of the peptide ratios delivers absolute quantification of the encoded peptides and in turn the target proteins for which they are surrogates. In this review, we discuss the criteria and difficulties associated with surrogate peptide selection and provide examples in the design of QconCATs for quantification of the proteins of the nuclear factor κB pathway.

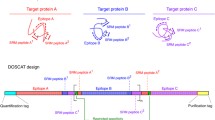
Workflow for QconCAT mediated protein quatification





Similar content being viewed by others
Abbreviations
- Glufib:
-
Glufibrinopeptide B
- GPMDB:
-
Global Proteome Machine Database
- GST:
-
Glutathione S-transferase
- His-tag:
-
Hexahistidine purification tag
- MALDI-ToF:
-
Matrix-assisted laser desorption/ionisation time of flight
- MRM:
-
Multiple reaction monitoring
- MS:
-
Mass spectrometry
- NF-κB:
-
Nuclear factor κB
- PrEST:
-
Protein epitope signature tag
- PSAQ:
-
Protein standards for absolute quantification
- QconCAT:
-
Quantification concatamer
- Rel:
-
Reticuloendotheliosis
- SDS-PAGE:
-
Sodium dodecyl sulphate polyacrylamide gel electrophoresis
- SILAC:
-
Stable isotope labelling by amino acids in cell culture
- SRM:
-
Selected reaction monitoring
References
Elliott MH, Smith DS, Parker CE, Borchers C (2009) J Mass Spectrom 44:1637–1660
Barnidge DR, Dratz EA, Martin T, Bonilla LE, Moran LB, Lindall A (2003) Anal Chem 75:445–451
Barr JR, Maggio VL, Patterson DGJ et al (1996) Clin Chem 42:1676–1682
Bettmer J (2010) Anal Bioanal Chem 397:3495–3502
Brun V, Masselon C, Garin J, Dupuis A (2009) J Proteomics 72:740–749
Kito K, Ito T (2008) Curr Genomics 9:263–274
Brun V, Dupuis A, Adrait A et al (2007) Mol Cell Proteomics 6:2139–2149
Dupuis A, Hennekinne JA, Garin J, Brun V (2008) Proteomics 8:4633–4636
Hennekinne JA, Brun V, De Buyser ML, Dupuis A, Ostyn A, Dragacci S (2009) Appl Environ Microbiol 75:882–884
Hanke S, Besir H, Oesterhelt D, Mann M (2008) J Proteome Res 7:1118–1130
Singh S, Springer M, Steen J, Kirschner MW, Steen H (2009) J Proteome Res 8:2201–2210
Zeiler M, Straube WL, Lundberg E, Uhlen M, Mann M (2012) Mol Cell Proteomics 11:O111.009613
Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP (2003) Proc Natl Acad Sci USA 100:6940–6945
Kirkpatrick DS, Gerber SA, Gygi SP (2005) Methods 35:265–273
Beynon RJ, Doherty MK, Pratt JM, Gaskell SJ (2005) Nat Methods 2:587–589
Pratt JM, Simpson DM, Doherty MK, Rivers J, Gaskell SJ, Beynon RJ (2006) Nat Protoc 1:1029–1043
Kito K, Ota K, Fujita T, Ito T (2007) J Proteome Res 6:792–800
Craig R, Cortens JP, Beavis RC (2004) J Proteome Res 3:1234–1242
Desiere F, Deutsch EW, Nesvizhskii AI et al (2005) Genome Biol 6:R9
Desiere F, Deutsch EW, King NL et al (2006) Nucleic Acids Res 34:D655–D658
Vizcaino JA, Cote R, Reisinger F et al (2009) Proteomics 9:4276–4283
Vizcaino JA, Reisinger F, Cote R, Martens L (2010) Curr Protoc Protein Sci Unit 25.4
Vizcaino JA, Reisinger F, Cote R, Martens L (2011) Methods Mol Biol 696:93–105
Brownridge P, Beynon RJ (2011) Methods 54:351–360
Eyers CE, Lawless C, Wedge DC, Lau KW, Gaskell SJ, Hubbard SJ (2011) Mol Cell Proteomics 10:M110.003384
Siepen JA, Keevil EJ, Knight D, Hubbard SJ (2007) J Proteome Res 6:399–408
Nanavati D, Gucek M, Milne JL, Subramaniam S, Markey SP (2008) Mol Cell Proteomics 7:442–447
Kudla G, Murray AW, Tollervey D, Plotkin JB (2009) Science 324:255–258
Brownridge P, Holman SW, Gaskell SJ et al (2011) Proteomics 11:2957–2970
Benita Y, Wise MJ, Lok MC, Humphery-Smith I, Oosting RS (2006) Mol Cell Proteomics 5:1567–1580
Ding C, Li Y, Kim BJ et al (2011) J Proteome Res 10:3652–3659
Mirzaei H, McBee JK, Watts J, Aebersold R (2008) Mol Cell Proteomics 7:813–823
Studier FW (1991) J Mol Biol 219:37–44
Brownridge P, Harman VM, Simpson DM, Beynon RJ (2012) Methods Mol Biol (in press)
Carroll KM, Simpson DM, Eyers CE et al (2011) Mol Cell Proteomics 10:M111.007633
Olinares PD, Kim J, Davis JI, van Wijk KJ (2011) Plant Cell 23:2348–2361
Hubbard SJ (1998) Biochim Biophys Acta 1382:191–206
Hubbard SJ, Beynon RJ, Thornton JM (1998) Protein Eng 11:349–359
Wu C, Robertson DH, Hubbard SJ, Gaskell SJ, Beynon RJ (1999) J Biol Chem 274:1108–1115
Moorthy AK, Savinova OV, Ho JQ, Wang VY, Vu D, Ghosh G (2006) EMBO J 25:1945–1956
Shih VF, Tsui R, Caldwell A, Hoffmann A (2011) Cell Res 21:86–102
Sun SC (2011) Cell Res 21:71–85
Mallick P, Schirle M, Chen SS et al (2007) Nat Biotechnol 25:125–131
Stergachis AB, MacLean B, Lee K, Stamatoyannopoulos JA, MacCoss MJ (2011) Nat Methods 8:1041–1043
Xiang Y, Remily-Wood ER, Oliveira V et al (2011) Mol Cell Proteomics 10:M110.005520
Eyers CE, Simpson DM, Wong SC, Beynon RJ, Gaskell SJ (2008) J Am Soc Mass Spectrom 19:1275–1280
Anderson L, Hunter CL (2006) Mol Cell Proteomics 5:573–588
Lebert D, Dupuis A, Garin J, Bruley C, Brun V (2011) Methods Mol Biol 753:93–115
Winter D, Seidler J, Kugelstadt D, Derrer B, Kappes B, Lehmann WD (2010) Proteomics 10:1510–1514
Zinn N, Winter D, Lehmann WD (2010) Anal Chem 82:2334–2340
Remily-Wood ER, Koomen JM (2012) J Mass Spectrom 47:188–194
Austin RJ, Chang DK, Holstein CA et al (2012) Proteomics. doi:10.1002/pmic.201100626
Bislev SL, Kusebauch U, Codrea MC et al (2012) J Proteome Res 11:1832–1843
Southworth PM, Hyde JE, Sims PF (2011) Malar J 10:315
Castro-Borges W, Simpson DM, Dowle A et al (2011) J Proteomics 74:1519–1533
Johnson H, Eyers CE, Eyers PA, Beynon RJ, Gaskell SJ (2009) J Am Soc Mass Spectrom 20:2211–2220
Rivers J, Hughes C, McKenna T et al (2011) PLoS One 6:e28902
Rivers J, Simpson DM, Robertson DH, Gaskell SJ, Beynon RJ (2007) Mol Cell Proteomics 6:1416–1427
Acknowledgments
D.M.S. and R.J.B. gratefully acknowledge funding from the Biotechnology and Biological Sciences Research Council (BBSRC) Systems Approach to Biological Research (SABR) grant BB/F005938/1 and BBSRC LOLA grant BB/G009112/1.
Author information
Authors and Affiliations
Corresponding author
Additional information
Published in the topical issue Quantitative Mass Spectrometry in Proteomics with guest editors Bernhard Küster and Marcus Bantscheff.
Rights and permissions
About this article
Cite this article
Simpson, D.M., Beynon, R.J. QconCATs: design and expression of concatenated protein standards for multiplexed protein quantification. Anal Bioanal Chem 404, 977–989 (2012). https://doi.org/10.1007/s00216-012-6230-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-012-6230-1