Abstract
The catalytic subunit of recombinant wild-type cyclic adenosine monophosphate-dependent protein kinase A (PKA) has been analyzed by a combination of 1D gel electrophoresis, in-gel digestion by trypsin, chymotrypsin, or endoproteinase AspN, and nano-ultraperformance liquid chromatography–MS/MS. The MS/MS spectra were annotated by MASCOT and the annotations were manually controlled. Using Ga(III)-immobilized metal ion affinity chromatography (IMAC), in addition to the four established autophosphorylation sites of the catalytic subunit of recombinant PKA, pSer10, pSer139, pThr197, and pSer338, six new phosphorylated residues have been characterized–pSer14, pThr48, pSer53, pSer212, pSer259, and pSer325. The established phosphorylation sites all are part of a PKA consensus motif and were found to be almost completely modified. In contrast, the newly detected sites were only partially phosphorylated. For estimation of their degree of phosphorylation, a method based on signal intensity measurements was used. For this purpose, signal intensities of all phospho- and non-phosphopeptides containing a particular site were added for estimation of site-specific phosphorylation degrees. This addition was performed over all peptides observed in the different digestion experiments, including their different charge states. pThr48 and pSer259 are located within PKA consensus motifs and were observed to be phosphorylated at 20% and 24%, respectively. pSer14 and pSer53 are located within inverted PKA consensus motifs and were found to be phosphorylated around 10% and 15%, respectively. The sequence environments of pSer212 and pSer325 have no similarity to the PKA consensus motif at all and were observed to be phosphorylated at about 5% or lower. All newly observed phosphorylation sites are located at the surface of the native protein structure of the PKA catalytic subunit. The results add new information on the theme of site-specific (auto)phosphorylation by PKA.

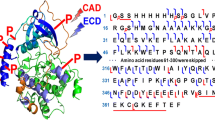
Sequence of protein kinase A Ca (P00517) with highlighted autophosphorylation sites as determined by UPLC-MS/MS. The 4 known autophosphorylation sites are printed in blue and the 6 newly detected autophosphorylation are printed in red.


Similar content being viewed by others
References
Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) The protein kinase complement of the human genome. Science 298:1912–1934
Walsh DA, Perkins JP, Krebs EG (1968) An Adenosine 3', 5'-monophosphate-dependent protein kinase from rabbit skeletal muscle. J Biol Chem 243:3763–3765
Taskén K, Aandahl EM (2004) Localized effects of cAMP mediated by distinct routes of protein kinase A. Physiol Rev 84:137–167
Gnad F, Ren SB, Cox J, Olsen JV, Macek B, Oroshi M, Mann M (2007) PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol 8:R250
Armstrong RN, Kondo H, Kaiser ET (1979) Cyclic AMP-dependent ATPase activity of bovine heart protein-kinase. Proc Natl Acad Sci USA 76:722–725
Cook PF, Neville ME, Vrana KE, Hartl FT, Roskoski R (1982) Adenosine cyclic 3', 5'-monophosphate dependent protein-kinase—kinetic mechanism for the bovine skeletal-muscle catalytic subunit. Biochemistry 21:5794–5799
Slice LW, Taylor SS (1989) Expression of the catalytic subunit of cAMP-dependent protein kinase in Escherichia coli. J Biol Chem 264:20940–20946
Uhler MD, Carmichael DF, Lee DC, Chrivia JC, Krebs EG, McKnight GS (1986) Isolation of cDNA clones coding for the catalytic subunit of mouse cAMP-dependent protein-kinase. Proc Natl Acad Sci USA 83:1300–1304
Moore MJ, Kanter JR, Jones KC, Taylor SS (2002) Phosphorylation of the catalytic subunit of protein kinase A—autophosphorylation versus phosphorylation by phosphoinositide-dependent kinase-1. J Biol Chem 277:47878–47884
Yonemoto W, McGlone ML, Grant B, Taylor SS (1997) Autophosphorylation of the catalytic subunit of cAMP-dependent protein kinase in Escherichia coli. Prot Eng 10:915–925
Jedrzejewski PT, Girod A, Tholey A, König N, Thullner S, Kinzel V, Bossemeyer D (1998) A conserved deamidation site at Asn 2 in the catalytic subunit of mammalian cAMP-dependent protein kinase detected by capillary LC-MS and tandem mass spectrometry. Prot Sci 7:457–469
Cheng XD, Ma YL, Moore M, Hemmings BA, Taylor SS (1998) Phosphorylation and activation of cAMP-dependent protein kinase by phosphoinositide-dependent protein kinase. Proc Natl Acad Sci USA 95:9849–9854
Herberg FW, Bell SM, Taylor SS (1993) Expression of the catalytic subunit of cAMP-dependent protein-kinase in Escherichia coli—multiple isozymes reflect different phosphorylation states. Prot Eng 6:771–777
Wind M, Edler M, Jakubowski N, Linscheid M, Wesch H, Lehmann WD (2001) Analysis of protein phosphorylation by capillary liquid chromatography coupled to element mass spectrometry with P-31 detection and to electrospray mass spectrometry. Anal Chem 73:29–35
Olsen SR, Uhler MD (1989) Affinity purification of the C-alpha and C-beta isoforms of the catalytic subunit of cAMP-dependent protein kinase. J Biol Chem 264:18662–18666
Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nature Prot 1:2856–2860
Winter D, Seidler J, Ziv Y, Shiloh Y, Lehmann WD (2009) Citrate boosts the performance of phosphopeptide analysis by UPLC-ESI-MS/MS. J Proteome Res 8:418–424
Winter D, Pipkorn R, Lehmann WD (2009) Separation of peptide isomers and confomers by ultra performance liquid chromatography. J Sep Sci 32:1111–1119
Gesellchen F, Bertinetti O, Herberg FW (2006) Analysis of posttranslational modifications exemplified using protein kinase A. Biochim Biophys Acta—Proteins and Proteomics 1764:1788–1800
Shen J, Smith RA, Stoll VS, Edalji R, Jakob C, Walter K, Gramling E, Dorwin S, Bartley D, Gunasekera A, Yang JG, Holzman T, Johnson RW (2004) Characterization of protein kinase A phosphorylation: multi-technique approach to phosphate mapping. Anal Biochem 324:204–218
Shoji S, Titani K, Demaille JG, Fischer EH (1979) Sequence of 2 phosphorylated sites in the catalytic subunit of bovine cardiac-muscle adenosine 3'-5'-monophosphate-dependent protein kinase. J Biol Chem 254:6211–6214
Grant BD, Hemmer W, Tsigelny I, Adams JA, Taylor SS (1998) Kinetic analyses of mutations in the glycine-rich loop of cAMP-dependent protein kinase. Biochemistry 37:7708–7715
Bartova I, Otyepka M, Kriz Z, Koca J (2004) Activation and inhibition of cyclin-dependent kinase-2 by phosphorylation; a molecular dynamics study reveals the functional importance of the glycine-rich loop. Prot Sci 13:1449–1457
Steen H, Jebanathirajah JA, Rush J, Morrice N, Kirschner MW (2006) Phosphorylation analysis by mass spectrometry—myths, facts, and the consequences for qualitative and quantitative measurements. Mol Cell Proteomics 5:172–181
Gropengiesser J, Varadarajan BT, Stephanowitz H, Krause E (2009) The relative influence of phosphorylation and methylation on responsiveness of peptides to MALDI and ESI mass spectrometry. J Mass Spectrom 44:821–831
Winter D, Kugelstadt D, Seidler J, Kappes B, Lehmann WD (2009) Protein phosphorylation influences proteolytic cleavage and kinase substrate properties exemplified by analysis of in vitro phosphorylated PfGAP45 by nanoUPLC-MS/MS. Anal Biochem Jun 20. [Epub ahead of print]
Thiede B, Lamer S, Mattow J, Siejak F, Dimmler C, Rudel T, Jungblut PR (2000) Analysis of missed cleavage sites, tryptophan oxidation and N-terminal pyroglutamylation after in-gel tryptic digestion. Rapid Commun Mass Spectrom 14:496–502
Tholey A, Pipkorn R, Bossemeyer D, Kinzel V, Reed J (2001) Influence of myristoylation, phosphorylation, and deamidation on the structural behavior of the N-terminus of the catalytic subunit of CAMP-dependent protein kinase. Biochemistry 40:225–231
Imanishi SY, Kochin V, Ferraris SE, de Thonel A, Pallari HM, Corthals GL, Eriksson JE (2007) Reference-facilitated phosphoproteomics - Fast and reliable phosphopeptide validation by microLC-ESI-Q-TOF MS/MS. Mol Cell Proteomics 6:1380–1391
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM1
(PDF 4571 kb)
Rights and permissions
About this article
Cite this article
Seidler, J., Adal, M., Kübler, D. et al. Analysis of autophosphorylation sites in the recombinant catalytic subunit alpha of cAMP-dependent kinase by nano-UPLC–ESI–MS/MS. Anal Bioanal Chem 395, 1713–1720 (2009). https://doi.org/10.1007/s00216-009-2932-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-009-2932-4