Abstract
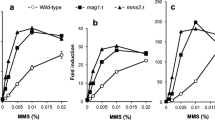
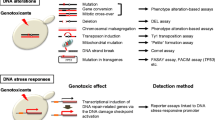
S. cerevisiae RNR3 and RAD54 gene transcription becomes strongly activated upon DNA damage. This property was used to construct yeast strains in which DNA damage can be monitored by a very sensitive fluorogenic assay in a convenient 96-well microtiter plate format. These strains carried stably integrated fusions of RNR3 or RAD54 promoters to the E. coli β-glucuronidase GUS gene. GUS activity was measured by fluorogenic detection, a method that greatly increases the precision and sensitivity of the assay. Detection levels were similar to those of real-time quantitative PCR methods and close to the limits of biological response. The two reporters differed in terms of fold-induction, activation kinetics, sensitivity and specificity upon exposure to a variety of genotoxic compounds. While RNR3-GUS showed the fastest response, RAD54-GUS showed the highest sensitivity: similar to previous reported sensitivities for bacterial and eukaryotic genotoxic detection systems. These reporter strains may complement current genotoxicity tests, but they also have the advantages of higher flexibility, requirement for shorter incubation times, and the capability of being fully automated. In addition, the intrinsic features of the system facilitate its easy improvement by genetic manipulating the yeast strain or by introducing mammalian metabolizing enzymes.




Similar content being viewed by others
References
Ames BN, Lee FD, Durston WE (1973) Proc Natl Acad Sci USA 70:782–786
Maron DM, Ames BN (1983) Mutat Res 113:173–215
Yasunaga K, Kiyonari A, Oikawa T, Abe N, Yoshikawa K (2004) Environ Mol Mutagen 44:329–345
Quillardet P, Huisman O, D’Ari R, Hofnung M (1982) Proc Natl Acad Sci USA 79:5971–5975
Jelinsky SA, Estep P, Church GM, Samson LD (2000) Mol Cell Biol 20:8157–8167
Friedberg EC, Siede W, Cooper AJ (1991) In: Jones EW, Pringle JR, Broach JR (eds) The molecular and cellular biology of the yeast Saccharomyces cerevisiae. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, Vol. 1, pp 147–192
Friedberg EC, Walker GC, Siede W (1995) DNA repair and mutagenesis. ASM, Washington, DC
Elledge SJ, Davis RW (1990) Genes Dev 4:740–751
Elledge SJ, Zhou Z, Allen, JB, Navas TA, Davis RW (1993) Bioessays 15:333–339
Jia X, Zhu Y, Xiao W (2002) Mutat Res 519:83–92
Jia X, Xiao W (2003) Toxicol Sci 75:82–88
Cole GM, Schild D, Lovett ST, Mortimer RK (1987) Mol Cell Biol 7:1078–1084
Gasch AP, Huang M, Metzner S, Botstein D, Elledge SJ, Brown PO (2001) Mol Biol Cell 12:2987–3003
Huang M, Zhou Z, Elledge SJ (1998) Cell 94:595–605
Walsh L, Schmuckli-Maurer J, Billinton N, Barker MG, Heyer WD, Walmsley RM (2002) Curr Genet 41:232–240
Afanassiev V, Sefton M, Anantachaiyong T, Barker G, Walmsley R, Wolfl S (2000) Mutat Res 464:297–308
Walmsley RM, Billinton N, Heyer WD (1997) Yeast 13:1535–1545
Billinton N, Barker MG, Michel CE, Knight AW, Heyer WD, Goddard NJ, Fielden PR, Walmsley RM (1998) Biosens Bioelectron 13:831–838
Knight AW, Goddard NJ, Billinton N, Cahill PA, Walmsley RM (2002) J Biochem Biophys Methods 51:165–177
Lichtenberg-Frate H, Schmitt M, Gellert G, Ludwig J (2003) Toxicol In Vitro 17:709–716
Cahill PA, Knight AW, Billinton N, Barker MG, Walsh L, Keenan PO, Williams, CV, Tweats DJ, Walmsley RM (2004) Mutagenesis 19:105–119
Knight AW, Keenan PO, Goddard NJ, Fielden PR, Walmsley RM (2004) J Environ Monit 6:71–79
Noguerol T, Boronat S, Jarque S, Barceló D, Piña B (2006) Talanta 69:358–359
Myung K, Kolodner RD (2003) DNA Repair (Amst) 2:243–258
Drablos F, Feyzi E, Aas PA, Vaagbo CB, Kavli B, Bratlie MS, Pena-Diaz J, Otterlei M, Slupphaug G, Krokan HE (2004) DNA Repair (Amst) 3:1389–1407
Thomas DC, Husain I, Chaney SG, Panigrahi GB, Walker IG (1991) Nucleic Acids Res 19:365–370
Sebastian J, Kraus B, Sancar GB (1990) Mol Cell Biol 10:4630–4637
Koc A, Wheeler LJ, Mathews CK, Merrill GF (2004) J Biol Chem 279:223–230
Jefferson RA, Kavanagh TA, Bevan MW (1987) Embo J 6:3901–3907
Goldstein AL, McCusker JH (1999) Yeast 15:1541–1553
Johnston M, Carlson M (1992) Gene expression. In: Jones EW, Pringle JR, Broach JR (eds) The molecular and cellular biology of the yeast Saccharomyces cerevisiae. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 193–282
Riu J, Marinez E, Barceló D, Ginebreda A, Tirapu LL (2001) Fresenius J Anal Chem 371:448–455
Farré M, Gonçalves C, Lacorte S, Barceló D, Alpendurada MF (2002) Anal Bioanal Chem 373:696–703
Routledge E, Sumpter J (1996) Environ Toxicol Chem 15:241–248
Garcia-Reyero N, Grau E, Castillo M, López de Alda MJ, Barceló D, Piña B (2001) Environ Toxicol Chem 20:1152–1158
Shetty RS, Deo SK, Liu Y, Daunert S (2004) Biotechnol Bioeng 88:664–670
Stocker J, Balluch D, Gsell M, Harms H, Feliciano J, Daunert S, Malik KA, Van der Meer JR (2003) Environ Sci Technol 37:4743–4750
Ramanathan S, Shi WP, Rosen BP, Daunert S (1998) Anal Chim Acta 369:189–195
Oda Y, Funasaka K, Kitano M, Nakama A, Yoshikura T (2004) Environ Mol Mutagen 43:10–19
Carro D, Bartra E, Piña B (2003) Appl Environ Microbiol 69:2161–2165
Acknowlegments
This work has been supported by the Spanish Ministry for Science and Technology (BIO2005-00840 and GEN2001-4707-C08-08). The contribution of the Centre de Referència en Biotecnologia de la Generalitat de Catalunya is also acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Boronat, S., Piña, B. Development of RNR3- and RAD54-GUS reporters for testing genotoxicity in Saccharomyces cerevisiae . Anal Bioanal Chem 386, 1625–1632 (2006). https://doi.org/10.1007/s00216-006-0751-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-006-0751-4