Abstract
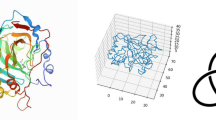
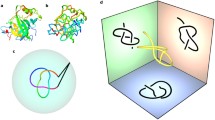
We present different means of classifying protein structure. One is made rigorous by mathematical knot invariants that coincide reasonably well with ordinary graphical fold classification and another classification is by packing analysis. Furthermore when constructing our mathematical fold classifications, we utilize standard neural network methods for predicting protein fold classes from amino acid sequences. We also make an analysis of the redundancy of the structural classifications in relation to function and ligand binding. Finally we advocate the use of combining the measurement of the VA, VCD, Raman, ROA, EA and ECD spectra with the primary sequence as a way to improve both the accuracy and reliability of fold class prediction schemes.
Similar content being viewed by others
References
Pascarella S and Argos P (1992). Protein Eng. 5: 121
Chothia C (1992). Nature 357: 543
Holm L and Sander C (1993). J Mol Biol 233: 123
Orengo CA, Flores TP, Taylor WR, Thornton JM (1993) Protein Engng 6:485
Jalkanen KJ and Suhai S (1996). Chem Phys 208: 81
Deng Z, Polavarapu PL, Ford SJ, Hecht L, Barron LD, Ewig CS and Jalkanen KJ (1996). J Phys Chem 100: 2025
Han W-G, Jalkanen KJ, Elstner M and Suhai S (1998). J Phys Chem B 102: 2587
Bohr H, Jalkanen KJ, Elstner M, Frimand K and Suhai S (1999). Chem Phys 246: 13
Tajkhorshid E, Jalkanen KJ and Suhai S (1998). J Phys Chem B 102: 5899
Frimand K, Bohr H, Jalkanen KJ and Suhai S (2000). Chem Phys 255: 165
Jalkanen KJ, Nieminen RM, Frimand K, Bohr J, Bohr H, Wade RC, Tajkhorshid E and Suhai S (2001). Chem Phys 265: 125
Knapp-Mohammady , Jalkanen KJ, Nardi F, Wade RC and Suhai S (1999). Chem Phys 240: 63
Jalkanen KJ,NieminenRM, Knapp- Mohammady, Suhai S (2003) 92:239
Jalkanen KJ (2003). J Phys Condens Matter 15: S1823
Jalkanen KJ, Elstner M and Suhai S (2004). J Mol Struct (Theochem) 675: 61
Bohr H, Frimand K, Jalkanen KJ, Nieminen RM and Suhai S (2001). Phys Rev E 64: 021905
Bohr H, Røgen P and Jalkanen KJ (2001). Comp Chem 26: 65
Callender RH, Dyer RB, Bilmanshin R and Woodruff WH (1998). Ann Rev Phys Chem 49: 173
Vanderkooi JM, Adar F and Erecinska M (1976). Eur J Biochem 64: 381
Nielsen KL, Idianai C, Henriksen A, Freis A, Beucucci M, Gajhede M, Smulevich G and Welinder KG (2001). Biochem 40: 11013
Smulevich G (1998). Biospectroscopy 4: S3
Kapetanaki S, Chouchase S, Girotto S, Yu S, Magliozzo RS and Schelvia JP (2003). Biochem 42: 385
Jones DJ, Taylor WR and Thornton JM (1992). Nature 358: 86
Reczko M and Bohr HG (1994). Nucleic Acids Res 22: 3616
Abola EE, Bernstein FC, Bryant SH, Koetzle TF, Weng J. (1987) Protein Data Bank. In: Crystallographic databases—information content, software systems, scientific Applications. In: Allen FH, Bergerhof G, Sievers R (eds.) 108–132, Data Commission of the international Union of Crystallography, Bonn/Cambridge/Chester.
Bassolino-Klimas D, Bruccoleri RE and Subramaniam S (1992). Protein Sci 1: 1465
Viswanathan M, Anchin JM, Droupadi PR, Mandal C, Linthicum DS, Subramaniam S (1994) Molecular Biophysics Technical Report UIUC-BI-MB-94-02
Goldstein RA, Luthey-Schulten ZA and Wolynes PG. (1992). Proc Natl Acad Sci USA 89: 9029
Ioerger TR, Rendell LA, Subramaniam S (1993) In: Proceedings of 1st International conference on intelligent systems formolecular biology, AAAI press, Menlo Park, pp 198–206
Bryant SH and Lawrence CE (1993). Proteins: Struct Func Genet 16: 92
Sippl MJ (1990). J Mol Biol 213: 859
Johnson MS, Overington JP and Blundell TL (1993). J Mol Biol 231: 735
Jones D and Thornton J (1993). J Comput Aided Mol Design 7: 439
Qian N and Sejnowski TJ (1988). J Mol Biol 202: 865
Bohr H, Bohr J, Brunak S, Cotterill RMJ, Lautrup B, Nørskov L, Olsen OH, Petersen SB (1988) FEBS Lett 241:223
Holley LH and Karplus M (1989). Proc Natl Acad Sci USA 86: 152
Bohr H, Bohr J, Brunak S, Cotterill RMJ, Fredholm H, Lautrup B and Petersen SB (1990). FEBS Lett 261: 43
Rumelhart DE, McClelland JL (eds.) (1986) Parallel distributed processing. MIT Press, Cambridge
Fahlman SE, Lebiere C (1990) In: Touretzky DS (ed.) Advances in neural information processing systems II, Morgan Kaufmann, Los Altos pp 524–532
Mathews BW (1975). Biochem Biophys Acta 405: 442
Stolorz P, Lapedes A, Yuan X (1991) Predicting protein secondary structure using neural net and statistical methods. (Los Alamos Preprint LA-UR-91-15)
Stolorz P, Lapedes A, Yuan X (1992) Predicting protein secondary structure using neural net and statistical methods. J Mol Biol 225:363
Zell A, Mache N, Sommer T, Korb T (1991) In: Proceedings of applications of neural networks conf., SPIE, Aerospace Sensing Intl. Symposium, Vol 1469 Orlando pp 708
Lesk AM (1991). “Protein architechture a practical approach”. Oxford University Press, Oxford
Walsh LL (1992) Protein Science 1:5, Diskette Appendix
Kauzmann W, Moore K and Schultz D (1974). Nature 248: 447
Reczko M, Bohr H (1994) In: Bohr H, Brunak S (eds.) Protein structure by distance analysis IOS Press, Amsterdam pp 87–97
Bohr H, Goldstein R, Wolynes PG (1992) AMSE Periodicals, Modelling, measurement and control C 31:55
Røgen P and Faen B (2003). Proc Nat Acad Sci (USA) 100: 119
Ashvar CS, Devlin FJ and Stephens PJ (1999). J Am Chem Soc 121: 2836
Hecht L and Barron LD (1994). J Raman Spectros 25: 443
Hecht L and Barron LD (1995). J Mol Struct 347: 449
Jalkanen KJ, Jürgensen VW and Degtyarenko IM (2005). Adv. Quan. Chem. 50: 91
Reczko M, Bohr H, Subramaniam S, Pamidighantam S, Hatzigeorgiou A (1994) In: Bohr H, Brunak S, Protein structure by distance analysis. IOS Press, Burke pp 277–286
Nielsen BG, Røgen P and Bohr H (2006). Math Comput Model 43: 401
Degtyarenko IM, Jalkanen KJ, Gurtovenko AA, Nieminen RM (2007) J Phys Chem B 111:4227
Beglov D and Roux B (1995). Biopolymers 35: 171
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ramnarayan, K., Bohr, H.G. & Jalkanen, K.J. Classification of protein fold classes by knot theory and prediction of folds by neural networks: A combined theoretical and experimental approach. Theor Chem Account 119, 265–274 (2008). https://doi.org/10.1007/s00214-007-0285-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00214-007-0285-7