Abstract
Commercially available antisera against five subtypes of muscarinic receptors and nine subtypes of adrenoceptors showed highly distinct immunohistochemical staining patterns in rat ureter and stomach. However, using the M1–4 muscarinic receptor subtypes and α2B-, β2-, and β3-adrenoceptors as examples, Western blots with membranes prepared from cell lines stably expressing various subtypes of muscarinic receptors or adrenoceptors revealed that each of the antisera recognized a set of proteins that differed between the cell lines used but lacked specificity for the claimed target receptor. We propose that receptor antibodies need better validation before they can reliably be used.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The use of antisera for immunohistochemical or other applications is based on the assumption that they exhibit high specificity for their target. Many commercially available antibodies are stated to be “affinity”-purified, but it is doubtful whether this purification procedure actually improves their specificity. Among the many criteria to evaluate specificity, a claim for selectivity of commercially available antisera is most often based upon a distinct immunohistochemical staining pattern, a single band of the expected size on Western blots, and/or the disappearance of staining after pre-absorption of the antiserum with purified epitope (cf. Petrusz et al. 1976 versus Swaab et al. 1977). However, all of these are soft criteria with justifiable exceptions. More meaningful “hard” specificity criteria include a staining pattern that is identical to that of an antibody raised against a different epitope on the same protein (Fischer et al. 2003) or the absence of staining in tissues of animals genetically deficient for the antigen (Swaab et al. 1977; Holmseth et al. 2006; Pradidarcheep et al. 2008). For example, we have recently reported that commercially available antisera against muscarinic receptor subtypes fulfilling at least one of the “soft” criteria lack specificity when tested in mice genetically deficient for the target receptor (Pradidarcheep et al. 2008). Therefore, additional validation of specificity of antibodies is often required.
While knockout animals are one of the most elegant ways to demonstrate specificity of a given antibody, their routine use for validation has several limitations. Firstly, knockout animals are not available, at least not for some researchers wishing to validate their antisera, for all target proteins. Secondly, an antibody for a given species of interest may not necessarily cross-react with its murine homolog. Finally, it is conceivable that a given antiserum may not be sufficiently specific for use, e.g., in immunohistochemistry but may nevertheless be suitable in Western blotting because band size can be used as an additional criterion to identify the protein of interest. An example of this is commercially available antibodies against Gs proteins which cross-react with smoothelin (Gsell et al. 2000); while this limits their use in immunohistochemistry, such antisera have nevertheless been helpful in studying regulation of Gs proteins by Western blots in many laboratories.
Therefore, we have tested whether genetically modified cell lines that stably express a receptor subtype of interest or related subtypes could be an attractive alternative for the use of knockout animals. Because all antisera against muscarinic receptors (MR) and adrenoceptors (AR) that were investigated failed this test and because this finding appears the rule rather than the exception, we propose to qualify antisera that meet the “hard” specificity criteria as “validated.”
Materials and methods
Antibodies
The antisera that were used are described in Table 1. All antibodies were designated affinity-purified preparations by the respective suppliers.
Animals
Wistar rats, 4–5 weeks old, were euthanized by instant decapitation under an O2/CO2 daze, in agreement with Dutch guidelines for experimental animals.
Immunohistochemical staining
The organs of the lower urinary and gastrointestinal tract were fixed overnight at 4°C by immersion in an ice-cold mixture of methanol–acetone–water (2:2:1; v/v). After dehydration and embedding, serial sections (7 μm) were mounted on poly-l-lysine-coated slides (for details, see Pradidarcheep et al. 2008). Antibody concentrations and staining times were chosen to assure a linear relation between antibody binding and staining intensity (van Straaten et al. 2006). Control sections, in which the primary antibody was omitted, were always included in the assays.
Cell lines expressing adrenergic or muscarinic receptors
Cell lines stably transfected with the human α1A-, α1B-, or α1D-AR (Rat-1 fibroblasts) or α2A-, α2B-, or α2C-AR (HEK-293 cells; Krege et al. 2000), β1-, β2-, or β3-AR (Chinese hamster ovary (CHO) cells; Niclauß et al. 2006), or M1-, M2-, M3-, or M4-MR (HEK-293 cells; Schmidt et al. 1995) were maintained as previously described. All cell lines had been tested for the expression of the transfected receptor subtype based on cell signaling responses or radioligand binding.
Western blotting
The preparation of whole-cell protein extracts in radioimmunoprecipitation assay buffer containing protease inhibitors, their separation on sodium dodecyl sulfate (SDS)-polyacrylamide gels, and blotting to polyvinylidene fluoride membranes (Millipore) were exactly as described (Pradidarcheep et al. 2008). After blocking, the membranes were exposed overnight to the antisera at the concentrations described in Table 1. Antibody binding was visualized with horseradish-peroxidase-conjugated goat antirabbit or goat antimouse secondary antibody and chemiluminescent imaging.
Results
Distinct staining patterns, including low background staining
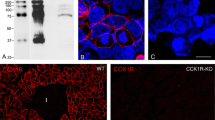
The staining patterns of five MR and nine AR subtype antibodies in the organs of the rat lower urinary tract, e.g., the ureter (Fig. 1), and the gastrointestinal tract, e.g., the stomach (Fig. 2), were distinct for each antiserum. Thus, the cellular staining patterns were different for all MR antisera. Although the α1A- and α1B-AR staining patterns of the ureter were similar, these antisera produced distinct staining patterns in the stomach. Conversely, the α1B-, α1D-, and α2A-AR antisera produced similar staining patterns in the stomach but distinct patterns in the ureter. These findings demonstrate that each of the antisera generated a staining pattern that differed from all other antisera, with little or no background noise. Unfortunately, such “pretty” staining patterns do not, as we will see, predict specificity for the cognate receptor.
Demonstration of lack of specificity
Western blots with extracts of organs of the lower urinary tract, gastrointestinal tract, and brain were prepared but the outcome was not satisfactory because all antisera yielded too many bands and did not allow specific detection of a major band likely to represent the target receptor (not shown). Whole-cell extracts of cell lines each stably expressing cloned human MR or AR subtypes were used for further evaluation of the various antisera. While, similar to the tissue sections, each MR antiserum generated a distinct “finger print” on the Western blot, that pattern was similar in cell lines expressing the cognate receptor subtype or related subtypes (Fig. 3). Similar observations were made with antisera supposedly acting on α2B-, β2-, or β3-AR (Fig. 4). These antisera, especially the α2B- and β3-AR preparations, produced a distinct banding pattern for Rat-1 fibroblasts (α1A-, α1B-, and α1D-AR lanes), HEK-293 cells (α2A-, α2B-, and α2C-AR lanes), and CHO cells (β1-, β2-, or β3-AR lanes) rather than a unique band (or set of bands in case posttranslational modifications had occurred) that was specific for the receptor that was transfected.
Staining pattern of antisera against MR subtypes on Western blots of cell lines each expressing one MR. Extracts (50-μg protein) of cell lines stably transfected with an expression vector encoding the MR indicated above each lane were electrophoresed, blotted, and stained for the presence of MR subtypes with the antisera. The calculated molecular weights for M1, M2, M3, and M4 receptors are 51, 52, 66, and 53 kDa, respectively. Note that each antiserum generated a unique staining pattern but that the pattern was identical irrespective of which MR subtype was expressed in the cell line
Staining pattern of antisera against AR subtypes on Western blots of cell lines each expressing one AR. Extracts (50-μg protein) of cell lines stably transfected with an expression vector encoding the AR indicated above each lane were electrophoresed, blotted, and stained for the presence of the α2B-, β2-, and β3-AR subtypes with the antisera. The calculated molecular weights for α1A-, α1B-, α1D-, α2A-, α2B-, α2C-, β1-, β2-, and β3-AR are 52, 56, 59, 50, 50, 50, 50, 47, and 43 kDa, respectively. Note that each antiserum shown generated a unique staining pattern but that none identified a band of the anticipated size and that many bands were shared irrespective of which AR subtype was expressed
Discussion
In our immunohistochemical experiments, all MR and AR antisera tested showed tissue- and antiserum-dependent staining patterns with a very satisfactory signal-to-background ratio in distinct tissues (Figs. 1 and 2). While each of the antisera exhibited a distinct pattern of bands for each host cell line, i.e., Rat-1 vs. CHO vs. HEK-293, in Western blots, none of them produced a distinct band in the cells expressing their target receptor as compared to those expressing related subtypes. While these findings raise questions about the selectivity of the antisera for their respective cognate receptors, several caveats need to be considered.
Firstly, it is possible that a given receptor presents differently in its native conformation (as seen in immunohistochemistry) as compared to the conformation adopted under denaturing condition (as seen on a SDS gel). While this means that an antibody lacking specificity on Western blots does not necessarily lack specificity in immunohistochemistry, our data nevertheless clearly do not support the idea of specificity. Moreover, transfected cells can of course also be used to test antibody specificity in immunocytochemistry. Secondly, our comparisons have not been based on non- or mock-transfected cells but rather on those transfected with related receptor subtypes. We actually consider this to be a strength of our approach as a given receptor upon overexpression may appear selective but loses that “selectivity” when studied at the lower expression levels occurring physiologically. This problem is circumvented by the use of the same cell line expressing related subtypes at a similar density. Thirdly, our immunoblots (based upon whole-cell extracts) show numerous bands for each antiserum, whereas those shown in a supplier’s catalog often show a single band only. While the number of bands due to low-affinity binding to other targets depends on the specific experimental conditions, any “specific” antibody should at least identify a unique major band even if the chosen conditions allow detection of various non-specific bands. However, that was not the case for any of the antibodies we have studied. Finally, the possibility should be considered that a given cell line has lost expression of the transfected receptor. While this is always a theoretical possibility, it is extremely unlikely that this applies to all 13 cell lines tested here. Moreover, we have verified the maintained presence of the transfected gene by cell signaling and/or radioligand binding studies for most cell lines within few months of the Western blot experiments.
In summary, we have shown that many receptor antibodies lack specificity for their cognate receptor in Western blots despite yielding very distinct staining patterns in tissues. We conclude that traditional criteria for antiserum specificity such as use of “pre-absorption,” elimination of staining by pre-incubation with the antigen, or the presence of only a single band on Western blots do not reliably predict specificity. While the use of corresponding knockout animals or the presence of identical staining patterns of antisera raised against distinct epitopes are the most reliable tests of specificity, we propose that the use of cell lines transfected with the target as compared to related receptors is a useful alternative to explore the specificity of antisera. We further propose that antisera, which pass one or both hard criteria and have this documented in the specification sheets, be identified as “validated” for easy recognition. It should be kept in mind, of course, that validation only assures the user that such an antiserum recognizes the macromolecule against which it was raised. Nevertheless, even a validated antiserum can, if applied improperly, still produce artifacts. The introduction of validated antisera could, nevertheless, provide the end user with a reliable starting preparation and, thus, avoid many fruitless and costly general confirmation assays that should have been carried out by the supplier. Finally, validated antisera would also further science by diminishing the scientific noise generated by erroneous data like those exposed in this series of articles.
Abbreviations
- MR:
-
muscarinic receptor
- AR:
-
adrenoceptor
References
Fischer DF, de Vos RAI, van Dijk R, de Vrij FMS, Proper EA, Sonnemans MAF, Verhage MC, Sluijs JA, Hobo B, Zouambia M, Jansen Steur ENH, Kamphorst W, Hol EM, van Leeuwen FW (2003) Disease-specific accumulation of mutant ubiquitin as a marker for proteasomal dysfunction in the brain. FASEB J 17:2014–2024
Gsell S, Eschenhagen T, Kaspareit G, Nose M, Scholz H, Behrens O, Wieland T (2000) Apparent up-regulation of stimulatory G-protein α subunits in the pregnant human myometrium is mimicked by elevated smoothelin expression. FASEB J 14:17–26
Holmseth S, Lehre KP, Danbolt NC (2006) Specificity controls for immunocytochemistry. Anat Embryol (Berl) 211:257–266
Krege S, Goepel M, Sperling H, Michel MC (2000) Affinity of trazodone at human penile α1- and α2-adrenoceptors. BJU Int 85:959–961
Niclauß N, Michel-Reher MB, Alewijnse AE, Michel MC (2006) Comparison of three radioligands for the labelling of human β-adrenoceptor subtypes. Naunyn-Schmiedeberg’s Arch Pharmacol 374:79–85
Petrusz P, Sar M, Ordronneau P, DiMeo P (1976) Specificity in immunocytochemical staining. J Histochem Cytochem 24:1110–1112
Pradidarcheep W, Labruyère WT, Dabhoiwala NF, Lamers WH (2008) Lack of specificity of commercially available antisera: better specifications needed. J Histochem Cytochem 56:1099–1111
Schmidt M, Bienek C, van Koppen CJ, Michel MC, Jakobs KH (1995) Differential calcium signalling by m2 and m3 muscarinic acetylcholine receptors in a single cell type. Naunyn-Schmiedeberg’s Arch Pharmacol 352:469–476
Swaab DF, Pool CW, Van Leeuwen FW (1977) Can specificity ever be proved in immunocytochemical staining? J Histochem Cytochem 25:388–391
van Straaten HW, He Y, van Duist MM, Labruyere WT, Vermeulen JL, van Dijk PJ, Ruijter JM, Lamers WH, Hakvoort TB (2006) Cellular concentrations of glutamine synthetase in murine organs. Biochem Cell Biol 84:215–231
Acknowledgements
This study was financed through a grant from the John L. Emmett Foundation for Urology, The Netherlands, and by research grants from the Thai government (#008/2550 and 125/2550).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Pradidarcheep, W., Stallen, J., Labruyère, W.T. et al. Lack of specificity of commercially available antisera against muscarinergic and adrenergic receptors. Naunyn-Schmied Arch Pharmacol 379, 397–402 (2009). https://doi.org/10.1007/s00210-009-0393-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00210-009-0393-0