Abstract
Two bacterial strains, 9H-EGSET and 15D-MOBT, were isolated from small freshwater habitats located near Salzburg, Austria. They showed the highest 16S rRNA sequence similarities of 100% and 99.9%, respectively, with type strains of species of the genus Aquirufa (Bacteroidota). Genome-based phylogenetic reconstructions with 119 amino acid sequences assigned the new taxa to the two distinct branches of the genus Aquirufa. Whole-genome average nucleotide identities were calculated with all possible pairs belonging to the genus. Values between 75.4% and 88.6% revealed that the two new strains represent each a new species. Like all, so far described members of the genus, they grew aerobically and chemoorganotrophically, were rod-shaped, red-pigmented, and motile by gliding, and showed genome sizes of about 3 Mbp and G + C values of about 40%. They could be distinguished by some phenotypic and chemotaxonomic features from their nearest related species. Until now, strain 9H-EGSET is the only one among the Aquirufa strains which contained traces of MK8 as respiratory quinone, and strain 15D-MOBT is the only one that formed tiny orange globules in liquid medium. The genome of strain 9H-EGSET comprised genes for the complete light-harvesting rhodopsin / retinal system, in the case of 15D-MOBT genes predicted for a nitrous oxide reductase were present. For the two new species of the genus Aquirufa, we propose to establish the names Aquirufa lenticrescens for strain 9H-EGSET (= JCM 34077 T = CIP 111926 T) and Aquirufa aurantiipilula for strain 15D-MOBT (= JCM 34078 T = CIP 111925 T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Aquirufa was first described by Pitt et al. in 2019 (Pitt et al. 2019) and comprises at the time of writing five species (Pitt et al. 2019, 2020; Sheu et al. 2020). The genera Aquirufa and Sandaracinomonas, described in 2020 (Chen et al. 2020), formed a lineage closely related to the genera Flectobacillus, Arcicella, and Pseudarcicella. The five genera of this clade were accommodated in the family Cytophagaceae (Skerman et al. 1980), belonging to the phylum Bacteroidota (Oren and Garrity 2021). Since genome-based phylogenetic analyses revealed that the family Cytophagaceae is non-monophyletic (Hahnke et al. 2016; García-López et al. 2019) García-López et al. proposed to split the Cytophagaceae into three distinct families. Besides Cytophagaceae and Flexibacteraceae, validated in 2020 (Oren and Garrity 2020b), they recommended using the validly published name Spirosomaceae proposed in 1978 (Larkin and Borrall 1978). At the time of establishment, the family included only the genera Spirosoma, Flectobacillus, and Runella (Larkin and Borrall 1978). In their emended description García-López et al. suggested adding 22 genera to the Spirosomaceae, this opinion was listed as a notification (Oren and Garrity 2020a) but is at the time of writing not yet validated. Nevertheless, while the three genera Flectobacillus, Arcicella, and Pseudarcicella were proposed for assignment to the Spirosomaceae, this would have to apply as well for the later described and rather closely related genera Aquirufa and Sandaracinomonas, which form a clade together with these three genera (Pitt et al. 2019).
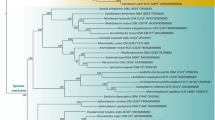
All by now, described members of this clade, as well as the new strains of this study, have in common that they were isolated from aquatic habitats or sources associated with water (Kämpfer et al. 2012; Chen et al. 2013, 2020; Sheu et al. 2017; Pitt et al. 2020). While the strains of the genera Aquirufa and Sandaracinomonas have genome sizes of about 3 Mbp, the available genome sequences of further species of the clade are double the size (Fig. 1).
Phylogenomic RAxML tree calculated with amino acid sequences obtained from 119 single-copy genes from all available genomes of the taxa from Fig. 4. Bar, 0.07 substitutions per nucleotide position
The genus Aquirufa is characterized by aerobic, chemoorganotrophic, rod-shaped, red-pigmented, and motile bacteria (Pitt et al. 2019). They seem to be widespread (Pitt et al. 2019) and sometimes abundant (Cruaud et al. 2020) and occur in a broad range of standing and running freshwater habitats (Cruaud et al. 2020; Pitt et al. 2019, 2020). Because the interspecific 16S rRNA gene sequence similarities of type strains are near 100% or exact 100%, the genus Aquirufa represents a cryptic species complex, which makes it impossible to assign publicly available 16S rRNA sequences of uncultured bacteria to particular Aquirufa species (Pitt et al. 2020). Nevertheless, there seemed to be an imbalance in the distribution of the members of the two main branches of the genus Aquirufa (Fig. 1). The NCBI Nucleotide BLAST website (Johnson et al. 2008) was used to search for matches for the 16S rRNA gene sequences similarities of ≥ 99.0%, considering isolates and environmental samples. A lot of detections affiliated with the ‘Aquirufa antheringensis branch’ (strain 30S-ANTBACT) were found in studies all over the world at various freshwater habitats. Some examples, which show the broad range of habitats are temperate rivers in Massachusetts (USA) (Crump and Hobbie 2005), Tibetan lakes (Zhang et al. 2013), Canadian permafrost thaw ponds (Rossi et al. 2013), freshwater lakes in Japan (Kojima et al. 2014), in Europe beside Austria, a mountain lake in France (Biderre-Petit et al. 2011), a drinking water reservoir in Greece (Lymperopoulou et al. 2012), and Karst cave water pools in Switzerland (Shabarova et al. 2013). Surprisingly, corresponding searches with the sequence of strain 59G-WUEMPELT (‘Aquirufa nivalisilvae branch’) revealed fewer detections, even though, this branch contains five species, while the ‘Aquirufa antheringensis branch’ comprises only two species, in both counting are the two new strains described here already included.
In this study, we present the description of two additional members of the genus Aquirufa. For this purpose, we sampled various freshwater habitats and inoculated agar plates with prefiltered water samples. We used the characteristic red pigmentation of the colonies to search for strains potentially belonging to the target group. Among the obtained strains were two, 9H-EGSET and 15D-MOBT, which each represent a new species for which we propose the names Aquirufa lenticrescens and Aquirufa aurantiipilula, respectively.
Materials and methods
Home habitats and isolation
Strain 9H-EGSET originated from Grossegelsee, a small natural pond located near the town of Mattsee, Austria. The water sample was taken from the lakeside in July 2019 at the approximate geographic coordinates 47.9622 N and 13.1245 E. Further sampling in July 2021 revealed pH 8.3 and conductivity of 471 µS cm−1. Strain 15D-MOBT originated from Moosbach, a small creek running through Mondsee, a village located near Salzburg, Austria. The water sample was taken in July 2019 at the approximate geographic coordinates 47.8616 N and 13.3399 E. Further sampling in July 2021 revealed pH 8.1 and conductivity of 619 µS cm−1. The isolation procedure aimed to obtain strains belonging to the genus Aquirufa. For that purpose, liquid nutrient broth, soytone, yeast extract (NSY) (Hahn et al. 2004) medium and NSY agar plates were used. The water sample of the pond Grossegelsee was filtered through a 0.65 µm pore size filter and the filtrate was subsequently spread on agar plates. The water sample of the creek Moosbach was treated in the same way, but using a 0.8 µm pore size filter. After one week of incubation at room temperature, small portions of colonies with the characteristic red pigmentation of Aquirufa strains were picked and transferred to 24-well plates filled with liquid NSY medium. These cultures were tested by partial sequencing of 5’-end of the 16S rRNA genes (primer pair 27F, 1492R) and comparative sequence analyses for affiliation with the genus Aquirufa. Promising cultures were subsequently purified. Because the 16S rRNA gene sequence similarities between Aquirufa species are relatively high (Pitt et al. 2020), the 16S rRNA gene is not appropriate to identify potential new species. Therefore, analyses of the sequences of the marker gene gyrB encoding the B subunit of the DNA gyrase (primer pair ArciLike-gyrB-F2 5’-TGCATTTCAAGCTATCTTGCC-3’ and ArciLike-gyrB-R2 5’-ACTTCATCTCCCATCAACATC-3’) were used to reduce the number of cultures to presumable candidates for new species. Finally, these remaining strains were genome-sequenced and the following analyses (see below) served to identify strains, which represent new species.
Phenotypic and chemotaxonomic characterization
The temperature range for growth was tested on a series of NSY agar plates exposed to increasing temperatures starting at 6 °C until no growth was observed. NaCl tolerance was tested using agar plates with various NaCl concentrations in 0.1% w/v steps. For testing anaerobic growth, an anaerobic chamber and standard NSY agar plates, as well as NSY plates supplemented with 2 g l−1 NaNO3, were used. For determination of cell morphology and cell dimension, well-growing liquid cultures were fixed with 2% paraformaldehyde, stained with DAPI (4′,6-diamidino-2-phenylindole), and investigated using an epifluorescence microscope (UV filter). Soft agar plates (1 g l−1 yeast extract, 0.1 g l−1 K2HPO4, and 2.0 g l−1 agar) were used to test the strain for motility (Hahn et al. 2017). One drop of a well-growing culture was placed in the center of these test plates, as well as on standard NSY plates, incubated at room temperature, and observed for several days. Assimilation of various substrates was tested using GEN III MicroPlatesT (Biolog), which detect utilization of substrates as electron donators by the subsequent reduction of a tetrazolium redox dye. Cells from well-growing liquid cultures were centrifuged and added to the inoculum medium so that the OD of the culture corresponded to 0.07 at a wavelength of 590 nm. After 48 h incubation at room temperature, the absorption was measured with a Multiskan FC (Thermo Scientific) at a wavelength of 595 nm. After subtracting the value of the negative control (without test substrate), obtained values from 0.016 to 0.05 were regarded as weak utilization, < 0.016 as negative, and > 0.05 as positive. The chemotaxonomic characterization included analyses of the composition of whole-cell fatty acids, polar lipids, and respiratory quinones. These analyses were carried out by the Identification Service, Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures GmbH. For all chemotaxonomic investigations, cells were inoculated into liquid NSY medium and harvested after 3 days of growth (room temperature) by centrifugation. For the whole-cell fatty acid composition, an Agilent Technologies 6890 N instrument and the Microbial Identification System (MIDI) Sherlock version 6.1 (method: TSBA6 database) were used as described by Sasser (Sasser 1990). The same extract was further analyzed by GC/MS to confirm peak identity and resolve summed features (Vieira et al. 2021). Double bond positions were analyzed by further derivatization to dimethyl disulfide adducts and subsequent GC/MS analysis (Moss and Lambert-Fair 1989). Polar lipids were extracted and analyzed as described by Tindall (Tindall 1990a, b) based on the method by Bligh and Dyer (Bligh and Dyer 1959). Polar lipids were separated by two-dimensional silica gel thin-layer chromatography. Total lipid material was detected using dodecamolybdophosphoric acid (Dmp) and specific functional groups by α-naphthol, ninhydrin, and molybdenum blue. Respiratory quinones were extracted by solid-phase extraction and analyzed by reversed-phase HPLC coupled to a diode array detector as described previously (Vieira et al. 2021).
Genomic characterization
DNA extraction and genome sequencing were performed as described previously (Hoetzinger et al. 2017). A shotgun library was paired-end sequenced on an Illumina HiSeq instrument (2 × 150 bp). De novo genome assembly was performed using the software SPAdes version 3.13.0 (Bankevich et al. 2012) and resulted in the case of strain 9H-EGSET in a closed genome with a coverage value of 690x, with a size of 2.5 Mbp and in the case of strain 15D-MOBT in 80 contigs with a coverage value of 500 × and a genome size of 3.1 Mbp. The obtained genome sequences were annotated by the NCBI Prokaryotic Genome Annotation Pipeline (Tatusova et al. 2016) and deposited at DDBJ/ENA/GenBank databases. For further comparative analyses, the genomes were also annotated by the Integrated Microbial Genomes & Microbiomes Expert Review (IMG/MER) system and incorporated into the IMG database (Chen et al. 2019). Homologous protein-encoding genes were identified using the IMG/MER (Chen et al. 2019) system (minimum 30% percentage identity, max. e-value 1e−5). The SEED viewer (Overbeek et al. 2005) was used for an amino acid sequence-based comparison of the new strains with their nearest relatives. Whole-genome average nucleotide identity (gANI) values and corresponding alignment fraction (AF) values were calculated for all possible pairs of the new strains with Aquirufa type strains using the IMG/MER system (Chen et al. 2019). In addition, digital DNA–DNA hybridization (dDDH) values were determined using the Type (Strain) Genome Server (Meier-Kolthoff et al. 2021).
Phylogenetic reconstructions
Phylogenetic trees were calculated using almost full-length sequences of the 16S rRNA gene and genome-based, using amino acid sequences of 119 single-copy marker genes (Parks et al. 2018). For the phylogenetic tree based on 16S rRNA gene sequences, the software MEGA X (Kumar et al. 2018) was used. The sequences were aligned and analyzed for the best-fitting substitution model. This resulted in neighbor-joining, maximum likelihood and parsimony reconstructions with the parameters Kimura 2 model (Kimura 1980), gamma-distributed (5 categories), invariant sites, and 1000 bootstrap replicates. For the phylogenetic tree based on genome sequences, 119 amino acid sequences of 120 protein-encoding genes recommended by Parks et al. (Parks et al. 2018) were used. One gene (protein familyTIGR0009) was not present in all considered genomes and was therefore omitted. The amino acid sequences were concatenated and aligned by MAFFT (Katoh et al. 2005). GBLOCKS (version 0.91b) (Castresana 2000) was used to filter out highly variable positions, which resulted in a reduction of the alignment from 47,304 to 42,980 positions in 310 selected blocks, which represents 90% of the alignment positions. A RAxML tree (Stamatakis 2014) with 100 bootstrap replicates was calculated using the CIPRES Science Gateway version 3.3 (Miller et al. 2010).
Results
Phenotypic and chemotaxonomic characteristics
Table 1 shows phenotypic and chemotaxonomic features of strains 9H-EGSET and 15D-MOBT and for comparison of the nearest related type strains of the genus Aquirufa. Strain 15D-MOBT showed a feature, which was by now unique among all strains of the genus. In liquid medium, the cells formed tiny orange globules (Fig. 2) and were not suspended, as usual for Aquirufa strains. This clump-forming made it impossible to get reliable results for the substrate (GEN III) tests and difficult to measure the cell dimensions. The substrate assimilation patterns can be found in Table 1, for not listed substrates, negative results were detected for all stains. Strain 9H-EGSET assimilated mucic acid beside Tween 40 and glucuronamide, which was not observed for the closely related strains of the genus Aquirufa. The patterns of the fatty acids were very similar to the patterns of the closely related type strains (Pitt et al. 2020) but differed by the absence of fatty acid Unknown 14.9591 (identified by mass spectrometry as C15:1ω4c) in the cells of strains 9H-EGSET and 15D-MOBT (Table S1, Online Resource 1 and Table 1). The major respiratory quinone was for both strains MK7, as so far detected for all Aquirufa strains, for strain 9H-EGSET traces of MK8 were identified in addition (Table 1). The results of analyses concerning the polar lipid patterns of strains 9H-EGSET and 15D-MOBT are given in Online Resource 1, Figure S1. For both strains, phosphatidylethanolamine was detected, as so far for all Aquirufa strains and occurred beside unidentified aminophospholipids and lipids, for strain 9H-EGSET the coloring with molybdenum blue revealed two additional aminophospholipids (Table 1).
Genomic characteristics
Some features of the genomes of strains 9H-EGSET and 15D-MOBT as well as of the type strains of all Aquirufa species and comparisons between them are listed in Table 2, genome sizes and G + C values can be found in Fig. 1. The sizes and G + C values of the genome sequences of the new strains consolidated the impression, that species of Aquirufa, which are closely related, have nearly the same genome sizes and G + C values (Fig. 1).
Analyses of the homologous protein-encoding genes (Table 2) showed that strain 9H-EGSET shared from its 2218 genes 90.7% with the type strain of the nearest relative Aquirufa antheringensis, strain 15D-MOBT from its 2674 genes as well around 90% with the type strains of the nearest related species A. beregesia, A. ecclesiirivi, and A. nivalisilvae. The amino acid sequence-based comparison of the new strains with their nearest relatives (Fig. 3) showed in case of strain 15D-MOBT similar patterns in each case of comparison with the three closest related type strains. Nevertheless, the comparisons illustrate genomic differentiation between the strains and their relatives, indicated by low sequence similarity regions and unique genes identifiable by white areas (no similarity) of the circles (Fig. 3). Regarding the specifically contained genes, strain 9H-EGSET showed genes predicted for synthesis of the complete light-harvesting rhodopsin / retinal system. Interestingly the strain had this feature in common with the type strain of the closest related species A. antheringensis, with a sequence similarity for the rhodopsin gene of 95.2% for nucleotides and 99.2% for amino acids. In contrast strain 15D-MOBT and the three related type strains of A. beregesia, A. ecclesiirivi and A. nivalisilvae lacked these genes (Table 2). Both new strains showed like all by now described strains of the genus Aquirufa several genes predicted for gliding motility associated proteins and lipoproteins and for cytochrome c oxidase as well as cytochrome cbb-3 oxidase. The new strains had genes presumably encoded for peroxidases comparable with the nearest related strains (Table 2). Table 2 shows as well the calculated gANI with the corresponding average AF values and dDDH percentages. Strain 9H-EGSET revealed the highest gANI value with the type strain of A. antheringensis with 88.6% (AF 87.9%) and the highest dDDH value amounted to 36.1% for the same pairing. Respective strain 15D-MOBT comparisons with the type strain of A. beregesia yielded the highest values, gANI 86.3% (AF 87.7) and a dDDH 31%.
Sequence-based comparison of the genomes of the two new strains with the genomes of the nearest related type strains of the genus Aquirufa. Left: Aquirufa lenticrescens 9H-EGSET (outer circle) and A. antheringensis 30S-ANTBACT (inner circle); right: A. aurantiipilula 15D-MOBT (in each case outer circle), A. ecclesiirivi 50A-KIRBAT (first inner circle), A. beregesia 50C-KIRBAT (second inner circle), A. nivalisilvae 59G-WUEMPELT (third inner circle). The colors indicate the percentage of the amino acid sequence identity for bidirectional and unidirectional best hits (see scale)
Phylogeny
Both phylogenetic trees (Figs. 1 and 4) showed nearly the same structure, only the position of Pseudarcicella hirudinis differed. In the genome-based phylogenetic tree, the cluster comprising the genera Flectobacillus, Arcicella, and Pseudarcicella with genome sizes around 6 Mbp was well separated from the cluster including the genera Sandaracinomonas and Aquirufa with significant smaller genome sizes of around 3 Mbp (Fig. 1). While the 16S rRNA sequence-based tree could not fully resolve the relationships between the species of the genus Aquirufa, the genome-based tree showed the phylogenetic distances of the two new strains to strains of the so far described species (Fig. 1).
Reconstruction of the phylogenetic position of the investigated strains based on almost full-length 16S rRNA gene sequences (1322 alignment positions). Shown is the neighbor-joining tree. Bootstrap values are shown from left to right for neighbor-joining, maximum likelihood, and maximum parsimony trees calculated with the same sequence set. Bar, 0.02 substitutions per nucleotide position; dots, nodes present in all calculated trees
Discussion
As mentioned above, the 16S rRNA sequence similarities between the species belonging to the genus Aquirufa are very high, so the new strain 9H-EGSET showed sequence similarity with A. antheringensis of 99.9% and strain 15D-MOBT of 100% with A. nivalisilvae and A. ecclesiirivi and 99.9% with A. beregesia. This circumstance of very high interspecific 16S rRNA gene similarity is well described for several genera (Stackebrandt and Ebers 2006; Hahn et al. 2016). Presumable are here the diversification rates, which lead to species separation higher than the substitution rates of the ribosomal genes. As evidence for a new species serves in this case the gANI or the dDDH value (Chun et al. 2018). For both new strains were the highest determined gANI values with 88.6% and 86.3%, respectively clearly under the accepted threshold of 95–96% utilized to delineate two prokaryotic species (Konstantinidis et al. 2006; Chun et al. 2018). The calculated dDDH values of maximal 36.1% and 31.0%, respectively, underlined this fact, as the accepted threshold is here 70% (Chun et al. 2018). So, strain 9H-EGSET and 15D-MOBT represent both new species. While most of the phenotypic and chemotaxonomic features of the two new strains resemble the patterns of the so far described Aquirufa species, some characteristics were suitable for discrimination (Table 1). Concerning the phenotypic features strain 9H-EGSET assimilated mucic acid beside Tween 40 and glucuronamide, which was not observed for the closely related strains of the genus Aquirufa (Table 1). Strain 15D-MOBT could be distinguished by the phenotypic feature, that cells formed tiny orange globules in liquid medium from all until now described strains of the genus Aquirufa and strain 9H-EGSET (Fig. 2 and Table 1). Concerning the chemotaxonomic traits, the cells of both new strains did not contain the fatty acid Unknown 14.959, as it was found for the closely related type strains as well the patterns of the polar lipids varied slightly (Table 1). Strain 9H-EGSET differed from all so far described type strains of the genus Aquirufa and strain 15D-MOBT by the occurrence of MK8 as respiratory quinone (Table 1) and (Sheu et al. 2020).
The genome-based phylogenetic tree (Fig. 1) in combination with the genomic data in Table 2 revealed some interesting points about the genus Aquirufa. The RAxML tree showed two distinct branches of the genus, in which the associated species had similar genome sizes and G + C values in common. This fact and comparisons with the branch length, thus the phylogenetic distances, between the genera Aquirufa and Sandaracinomonas on the one hand and Flectobacillus, Arcicella, and Pseudarcicella, on the other hand, suggested that the two main branches of the current genus Aquirufa could be regarded as two separated genera. The relatively low AF values of around 35% for genome pairs composed of strains from both Aquirufa branches supported this impression. On the other hand, the phenotypic and chemotaxonomic properties of the strains related to the two branches were very similar. Since, besides the 16S rRNA gene similarity, which is here not suitable, clear thresholds for separating genera are lacking this point must be left open. Maybe further isolations of new strains will enhance the impression of two distinct genera or will reveal ‘missing links’ between the two Aquirufa branches or between the genera Aquirufa and Sandaracinomonas. Therefore, we propose here only the establishment of two new species of the genus Aquirufa, with the name Aquirufa lenticrescens for strain 9H-EGSET and Aquirufa aurantiipilula for strain 15D-MOBT.
The occurrence of genes predicted for oxidases and peroxidases suggested that strains 9H-EGSET and 15D-MOBT occur under aerobic conditions, especially the cytochrome cbb-3 oxidase indicated tolerance to microaerobic conditions (Pitcher et al. 2002). Genes putatively encoding for synthesis of the complete light-harvesting rhodopsin / retinal system in strain 9H-EGSET suggested a distribution in upper water layers, where enough light is available. The gliding motility, observed in experiments and confirmed by the occurrence of corresponding genes, implicated the association with substrate.
Description of Aquirufa lenticrescens sp. nov. (len.ti.cres’cens. L. masc. adj. lentus, slow; L. pres. part. crescens, growing; N.L. part. adj. lenticrescens, slow-growing)
Cells form rods, about 1.2 µm long and 0.5 µm wide. Colonies grown on NSY or R2A agar are bright red, in older stages dark red, pigmented, circular, and convex with a smooth surface. Liquid cultures grown in NSY or R2A medium have a red–orange coloring. Cells can move on soft agar. Growth occurs at 5–31 °C and in 0–0.1% (w) NaCl. Cells assimilate Tween 40, weakly assimilate mucic acid and glucuronamide, they do not assimilate the rest of the GEN III MicroPlatesT (Biolog). Major fatty acids are iso-C15:0, anteiso-C15:0, and summed feature 3 (C16:1ω7c / C16:1ω6c, identified by mass spectrometry as C16:1ω7c). Polar lipids are phosphatidylethanolamine, three unidentified aminophospholipid, and two unidentified polar lipids. Major respiratory quinone is MK7, traces of MK8 occur. The genome of the type strain is characterized by a size of 2.5 Mbp and a G + C content of 42.2 mol%.
The type strain is 9H-EGSET (= JCM 34077 T = CIP 111926 T), which was isolated from Grossegelsee, a small pond, located near the town of Mattsee, Austria.
The accession number of the genome sequence is CP049834 and MZ665465 of the 16S rRNA sequence.
Description of Aquirufa aurantiipilula sp. nov. (au.ran.ti.i.pi’lu.la. L. masc. adj. aurantius, orange; L. fem. n. pilula, small ball; N.L. fem. n. aurantiipilula, orange colored small ball)
Cells form rods, about 1.9 µm long and 0.6 µm wide. Colonies grown on NSY or R2A agar are bright red, in older stages dark red, pigmented, circular, and convex with a smooth surface. Grown in liquid NSY or R2A medium cells accumulate to tiny orange globules. Cells can move on soft agar. Growth occurs at 5–35 °C and in 0–0.3% (w) NaCl. Major fatty acids are iso-C15:0, anteiso-C15:0, and summed feature 3 (C16:1ω7c / C16:1ω6c, identified by mass spectrometry as C16:1ω7c). Polar lipids are phosphatidylethanolamine, three unidentified aminophospholipid, and two unidentified polar lipids. Major respiratory quinone is MK7. The genome of the type strain is characterized by a size of 3.1 Mbp and a G + C content of 38.0 mol%.
The type strain is 15D-MOBT (= JCM 34078 T = CIP 111925 T), which was isolated from Moosbach, a small creek, running through the town Mondsee, near Salzburg, Austria.
The accession number of the genome sequence is JAANOG000000000 and MZ665466 of the 16S rRNA sequence.
Data availability
The sequences were available at DDBJ/ENA/GenBank. The accession numbers of the whole-genome sequence of strain 9H-EGSET are CP049834 and of strain 15D-MOBT JAANOG000000000 of the 16S rRNA gene sequence of strain 9H-EGSET is MZ665465 and of strain 15D-MOBT MZ665466. All genomes in Table 2 are available in the IMG/MER system (ID numbers see Table 2).
Abbreviations
- NSY medium:
-
Nutrient broth, soytone, yeast extract medium
- IMG/MER:
-
Integrated Microbial Genomes and Microbiomes Expert Review
- gANI:
-
Whole-genome average nucleotide identity
- AF:
-
Alignment fraction
- dDDH:
-
Digital DNA–DNA hybridization
- A :
-
Aquirufa
References
Bankevich A, Nurk S, Antipov D et al (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. https://doi.org/10.1089/cmb.2012.0021
Biderre-Petit C, Boucher D, Kuever J et al (2011) Identification of sulfur-cycle prokaryotes in a low-sulfate lake (Lake Pavin) using aprA and 16S rRNA gene markers. Microb Ecol 61:313–327. https://doi.org/10.1007/s00248-010-9769-4
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem 37:911–917
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. https://doi.org/10.1093/oxfordjournals.molbev.a026334
Chen WM, Yang SH, Young CC, Sheu SY (2013) Arcicella rigui sp. nov., isolated from water of a wetland, and emended descriptions of the genus Arcicella, Arcicella aquatica, Arcicella rosea and Arcicella aurantiaca. Int J Syst Evol Microbiol 63:134–140. https://doi.org/10.1099/ijs.0.037242-0
Chen IMA, Chu K, Palaniappan K et al (2019) IMG/M vol 5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res 47:D666–D677. https://doi.org/10.1093/nar/gky901
Chen W-M, Cai C-Y, Sheu S-Y (2020) Sandaracinomonas limnophila gen. nov., sp. nov., a new member of the family Cytophagaceae isolated from a freshwater mesocosm. Int J Syst Evol Microbiol 70:2178–2185. https://doi.org/10.1099/ijsem.0.003532
Chun J, Oren A, Ventosa A et al (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466. https://doi.org/10.1099/ijsem.0.002516
Cruaud P, Vigneron A, Dorea CC et al (2020) Rapid changes in microbial community structures along a meandering River. Microorganisms 8(11):1631. https://doi.org/10.3390/microorganisms8111631
Crump BC, Hobbie JE (2005) Synchrony and seasonality in bacterioplankton communities of two temperate rivers. Limnol Oceanogr 50:1718–1729. https://doi.org/10.4319/lo.2005.50.6.1718
García-López M, Meier-Kolthoff JP, Tindall BJ et al (2019) Analysis of 1000 type-strain genomes improves taxonomic classification of Bacteroidetes. Front Microbiol. https://doi.org/10.3389/fmicb.2019.02083
Hahn MW, Stadler P, Wu QL, Pöckl M (2004) The filtration-acclimatization method for isolation of an important fraction of the not readily cultivable bacteria. J Microbiol Methods 57:379–390. https://doi.org/10.1016/j.mimet.2004.02.004
Hahn MW, Jezberova J, Koll U, Saueressig-Beck T, Schmidt J (2016) Complete ecological isolation and cryptic diversity in Polynucleobacter bacteria not resolved by 16S rRNA gene sequences. ISME J 10:1642–1655. https://doi.org/10.1038/ismej.2015.237
Hahn MW, Schmidt J, Koll U et al (2017) Silvanigrella aquatica gen. nov., sp. nov., isolated from a freshwater lake, description of Silvanigrellaceae fam. nov. and Silvanigrellales ord. nov., reclassification of the order Bdellovibrionales in the class Oligoflexia, reclassification of the families Bacteriovoracaceae and Halobacteriovoraceae in the new order Bacteriovoracales ord. nov., and reclassification of the family Pseudobacteriovoracaceae in the order Oligoflexales. Int J Syst Evol Microbiol 67:2555–2568. https://doi.org/10.1099/ijsem.0.001965
Hahnke RL, Meier-Kolthoff JP, García-López M et al (2016) Genome-based taxonomic classification of Bacteroidetes. Front Microbiol 7:2003. https://doi.org/10.3389/fmicb.2016.02003
Hoetzinger M, Schmidt J, Jezberova J, Koll U, Hahn MW (2017) Microdiversification of a pelagic Polynucleobacter species is mainly driven by acquisition of genomic islands from a partially interspecific gene pool. Appl Environ Microbiol 83:19. https://doi.org/10.1128/aem.02266-16
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5-9. https://doi.org/10.1093/nar/gkn201
Kämpfer P, Busse HJ, Longaric I, Rossello-Mora R, Galatis H, Lodders N (2012) Pseudarcicella hirudinis gen. nov., sp. nov., isolated from the skin of the medical leech Hirudo medicinalis. Int J Syst Evol Microbiol 62:2247–2251. https://doi.org/10.1099/ijs.0.037390-0
Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518. https://doi.org/10.1093/nar/gki198
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide-sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/bf01731581
Kojima H, Watanabe T, Iwata T, Fukui M (2014) Identification of major planktonic sulfur oxidizers in stratified freshwater lake. PLoS One. https://doi.org/10.1371/journal.pone.0093877
Konstantinidis KT, Ramette A, Tiedje JM (2006) The bacterial species definition in the genomic era. Phil Trans R Soc B-Biol Sci 361:1929–1940. https://doi.org/10.1098/rstb.2006.1920
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbey/msy096
Larkin JM, Borrall R (1978) Spirosomaceae, a new family to contain the genera Spirosoma Migula 1894, Flectobacillus Larkin et al. 1977 and Runella Larkin and Williams 1978. Int J Syst Evol Microbiol 28:595–596. https://doi.org/10.1099/00207713-28-4-595
Lymperopoulou DS, Kormas KA, Karagouni AD (2012) Variability of prokaryotic community structure in a drinking water reservoir (Marathonas, Greece). Microbes Environ 27:1–8. https://doi.org/10.1264/jsme2.me11253
Meier-Kolthoff JP, Carbasse JS, Peinado-Olarte RL, Göker M (2021) TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res 50:D801–D807. https://doi.org/10.1093/nar/gkab902
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the gateway computing environments workshop (GCE). IEEE, New Orleans, LA, pp 1–8
Moss CW, Lambert-Fair MA (1989) Location of double bonds in monounsaturated fatty acids of Campylobacter cryaerophila with dimethyl disulfide derivatives and combined gas chromatography-mass spectrometry. J Clin Microbiol 27:1467–1470. https://doi.org/10.1128/jcm.27.7.1467-1470.1989
Oren A, Garrity G (2020a) Notification of changes in taxonomic opinion previously published outside the IJSEM. Int J Syst Evol Microbiol 70:4061–4090. https://doi.org/10.1099/ijsem.0.004245
Oren A, Garrity GM (2020b) List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol 70:2960–2966. https://doi.org/10.1099/ijsem.0.004156
Oren A, Garrity GM (2021) Valid publication of the names of forty-two phyla of prokaryotes. Int J Syst Evol Microbiol. https://doi.org/10.1099/ijsem.0.005056
Overbeek R, Begley T, Butler RM et al (2005) The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33:5691–5702. https://doi.org/10.1093/nar/gki866
Parks DH, Chuvochina M, Waite DW et al (2018) A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36:996–1004. https://doi.org/10.1038/nbt.4229
Pitcher RS, Brittain T, Watmugh NJ (2002) Cytochrome cbb3 oxidase and bacterial microaerobic metabolism. Biochem Soc Trans 30:653–658. https://doi.org/10.1042/bst0300653
Pitt A, Schmidt J, Koll U, Hahn MW (2019) Aquirufa antheringensis gen. nov., sp. nov. and Aquirufa nivalisilvae sp. nov., representing a new genus of widespread freshwater bacteria. Int J Syst Evol Microbiol 69:2739–2749. https://doi.org/10.1099/ijsem.0.003554
Pitt A, Koll U, Schmidt J, Hahn MW (2020) Aquirufa ecclesiirivi sp. nov. and Aquirufa beregesia sp. nov., isolated from a small creek and classification of Allopseudarcicella aquatilis as a later heterotypic synonym of Aquirufa nivalisilvae. Int J Syst Evol Microbiol 70:4602–4609. https://doi.org/10.1099/ijsem.0.004319
Rossi PG, Laurion I, Lovejoy C (2013) Distribution and identity of bacteria in subarctic permafrost thaw ponds. Aquat Microb Ecol 69:231–245
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. USFCC Newsl 20:16
Shabarova T, Widmer F, Pernthaler J (2013) Mass effects meet species sorting: transformations of microbial assemblages in epiphreatic subsurface karst water pools. Environ Microbiol 15:2476–2488. https://doi.org/10.1111/1462-2920.12124
Sheu SY, Liu LP, Chen WM (2017) Flectobacillus pallidus sp. nov., isolated from a freshwater spring. Int J Syst Evol Microbiol 67:1126–1132. https://doi.org/10.1099/ijsem.0.001774
Sheu S-Y, Chen T-Y, Chen W-M (2020) Aquirufa rosea sp. nov., isolated from a freshwater lake. Int J Syst Evol Microbiol 70:3145–3153. https://doi.org/10.1099/ijsem.0.004147
Skerman VBD, McGowan V, Sneath PHA (1980) Approved lists of bacterial names. Int J Syst Evol Microbiol 30:225–420. https://doi.org/10.1099/00207713-30-1-225
Stackebrandt E, Ebers J (2006) Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 8:6–9
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Tatusova T, DiCuccio M, Badretdin A et al (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. https://doi.org/10.1093/nar/gkw569
Tindall BJ (1990a) A comparative study of the lipid composition of Halobacterium saccharovorum from various sources. Syst Appl Microbiol 13:128–130. https://doi.org/10.1016/s0723-2020(11)80158-x
Tindall BJ (1990b) Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol Lett 66:199–202
Vieira S, Huber KJ, Neumann-Schaal M et al (2021) Usitatibacter rugosus gen. nov., sp. nov. and Usitatibacter palustris sp. nov., novel members of Usitatibacteraceae fam. nov. within the order Nitrosomonadales isolated from soil. Int J Syst Evol Microbiol. https://doi.org/10.1099/ijsem.0.004631
Zhang R, Wu Q, Piceno YM et al (2013) Diversity of bacterioplankton in contrasting Tibetan lakes revealed by high-density microarray and clone library analysis. FEMS Microbiol Ecol 86:277–287. https://doi.org/10.1111/1574-6941.12160
Acknowledgements
We are grateful to Ingrid Lugstein and Noah Bruckner for their assistance in water sampling and laboratory work. We are also grateful to Jana Mierl and Lea Huemer for assistance in water sampling and data analyses and to Birgit Grün, Anika Wasner, Anja Frühling and Gesa Martens for technical assistance in chemotaxonmic analyses.
Funding
Open access funding provided by University of Innsbruck and Medical University of Innsbruck. This study was supported by ‘Sparkling Science’ a research program of the Austrian Federal Ministry of Education, Science and Research (BMBWF) project SPA 06/065, and the program ‘Talente’ of the Austrian Research Promotion Agency (FFG).
Author information
Authors and Affiliations
Contributions
UK, JS, and SK isolated the strains. All authors contributed to the investigations. AP analysed the data and wrote the manuscript. MWH, MNS, and JW provided comments on the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose. The authors declare no competing interests.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pitt, A., Koll, U., Schmidt, J. et al. Aquirufa lenticrescens sp. nov. and Aquirufa aurantiipilula sp. nov.: two new species of a lineage of widespread freshwater bacteria. Arch Microbiol 204, 356 (2022). https://doi.org/10.1007/s00203-022-02950-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00203-022-02950-6