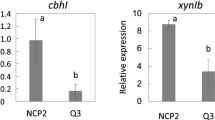
Abstract
Cellulomonas flavigena produces a battery of cellulase components that act concertedly to degrade cellulose. The addition of cAMP to repressed C. flavigena cultures released catabolic repression, while addition of cAMP to induced C. flavigena cultures led to a cellobiohydrolase hyperproduction. Exogenous cAMP showed positive regulation on cellobiohydrolase production in C. flavigena grown on sugar cane bagasse. A C. flavigena cellobiohydrolase gene was cloned (named celA), which coded for a 71- kDa enzyme. Upstream, a repressor celR1, identified as a 38 kDa protein, was monitored by use of polyclonal antibodies.



Similar content being viewed by others
References
Avitia CI, Castellanos-Juárez FX, Sánchez E, Téllez-Valencia A, Fajardo-Cavazos P, Nicholson WL, Pedraza-Reyes M (2000) Temporal secretion of multicellulolytic system in Myxobacter sp. AL-1. Molecular cloning and heterologous expression of cel9 encoding a modular endocellulase clustered in an operon with cel 48, an exocellobiohydrolase gene. Eur J Biochem 267:7058–7064
Bagga PS, Sandhu DK, Sharma S (1991) Effect of exogenous cyclic AMP on catabolic repression of cellulases formation in Aspergillus nidulans. Acta Biotechnol 11:395–402
Bayer EA, Chanzy H, Lamed R, Shoham Y (1998) Cellulose, cellulases and cellulosomes. Curr Op Struct Biol 8:548–557
Botsford JL, Harman JG (1992) Cyclic AMP in prokaryotes. Microbiol Rev 56:100–122
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72:248–254
Demain AL, Newcomb M, Wu JHD (2005) Cellulase, clostridia, and ethanol. Microbiol Mol Biol Rev 69:124–154
Desphande MV, Eriksson KE, Pettersson LG (1984) An assay for selective determination of exo-1, 4,-β-glucanases in a mixture of cellulolytic enzymes. Anal Biochem 138:481–487
Emory S, Bouvet P, Belasco J (1992) A 5′terminal stem-loop structure can stabilize mRNA in Escherichia coli. Gene Dev 6:135–148
Gilkes NR, Henrissat B, Kilburn DG, Miller RC Jr, Warren RAJ (1991) Domains in microbial-1, 4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev 55:303–315
Gutiérrez-Nava A, Herrera-Herrera JA, Mayorga-Reyes L, Salgado LM, Ponce-Noyola T (2003) Expression and characterization of the celcflB gene from Cellulomonas flavigena encoding an endo-beta-1, 4-glucanase. Curr Microbiol 47:359–363
Henrissat B, Davies G (1997) Structural and sequence-based classification of glycoside hydrolases. Curr Opin Struct Biol 7:637–644
Juhász T, Szengyel Z, Réczey K, Siika-Aho M, Viikari L (2005) Characterization of cellulases and hemicellulases produced by Trichoderma reesei on various carbon sources. Process Biochem 40:3519–3525
Kim JO, Park SR, Lim W, Ryu SK, Kim M, Ch An, Cho SJ, Park Y, Kim J, Yun H (2000) Cloning and characterization of thermostable endoglucanase (Cel8Y) from the hyperthermophilic Aquifex aeolicus VF5. Biochem Biophys Res Comm 279:420–426
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the folin phenol reagent. J Biol Chem 193:265–275
Mayorga Reyes L, Gutiérrez-Nava A, Salgado L, Ponce-Noyola T (2005) Aislamiento de una clona que contiene un gen de xilanasa a partir de una genoteca de Cellulomonas flavigena. Rev Mex C Farm 36:5–9
Meinke A, Gilkes NR, Kwan E, Kilburn DG, Warren ARJ, Miller RC Jr (1994) Cellobiohydrolase A (CbhA) from the cellulolytic bacterium Cellulomonas fimi is a ß-1, 4-exocellobiohydrolase analogous to Trichoderma reesei CBHII. Mol Microbiol 12:413–422
Moser B, Gilkes NR, Kilburn DG, Warren RAJ, Miller RC Jr (1989) Purification and characterization of endoglucanase C of Cellulomonas fimi, cloning of the gene, and analysis of in vivo transcripts of the gene. Appl Environ Microbiol 55:2480–2487
Nguyen CC, Saier MH Jr (1995) Phylogenetic, structural and functional analyses of the LacI-GalR family of bacterial transcription factors. FEBS Lett 18:98–102
Ponce-Noyola T, de la Torre M (2001) Regulation of cellulases and xylanases from derepressed mutant of Cellulomonas flavigena growing on sugar cane bagasse in continuous culture. Biores Technol 78:285–291
Rabinovich ML, Melnick MS, Bolobova AV (2002a) Microbial cellulases (Review). Appl Biochem Microbio 38:305–321
Rabinovich ML, Melnick MS, Bolobova AV (2002b) The structure and mechanism of action of cellulolytic enzymes. Biochemistry 8:850–871
Sambrook JE, Fritsche F, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Press, Cold Spring Harbor
Sánchez-Herrera LM, Ramos-Valdivia AC, de la Torre M, Salgado LM, Ponce-Noyola T (2007) Differential expression of cellulases and xylanases by Cellulomonas flavigena grown on different carbon sources. Appl Microbiol Biotechnol 77:589–595
Schwarz WH (2001) The cellulosome and cellulose degradation by anaerobic bacteria. App Microbiol Biotechnol 56:634–649
Shulein M (2000) Protein engineering of cellulases. Biochim Biophys Acta 1543:239–252
Spirindov NA, Wilson DB (1999) Characterization and cloning of CelR, a transcriptional regulator of cellulase genes from Thermomonospora fusca. J Biol Chem 274:13127–13132
Voet D, Voet JG (2003) Biochemistry. Wiley, New York
Wilson DB (2004) Studies of the Thermobifida fusca plant cell wall degrading enzymes. Chem Record 4:72–82
Wood TM, Garcia-Campayo V (1990) Enzymology of cellulose degradation. Biodegradation 1:147–161
Wood WE, Neubauer DG, Stutzenberger FJ (1984) Cyclic AMP levels during induction and repression of cellulase biosynthesis in Thermomonospora curvata. J Bacteriol 160:1047–1054
Acknowledgments
This work was supported by grant 45678-Z from Consejo Nacional de Ciencia y Tecnología (Conacyt), Mexico. JAHH was supported by PhD fellowship from Conacyt. The authors thank M. Mercado-Morales for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Jorge Membrillo-Hernández.
Rights and permissions
About this article
Cite this article
Herrera-Herrera, J.A., Pérez-Avalos, O., Salgado, L.M. et al. Cyclic AMP regulates the biosynthesis of cellobiohydrolase in Cellulomonas flavigena growing in sugar cane bagasse. Arch Microbiol 191, 745–750 (2009). https://doi.org/10.1007/s00203-009-0502-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-009-0502-y