Abstract
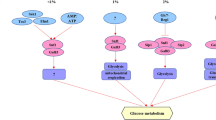
In yeasts, several sensing systems localized to the plasma membrane which transduce information regarding the availability and quality of nitrogen and carbon sources and work in parallel with the intracellular nutrient-sensing systems, regulate the expression and activity of proteins involved in nutrient uptake and utilization. The aim of this work was to establish whether the cellular signals triggered by amino acids modify the expression of the UGA4 gene which encodes the δ-aminolevulinic (ALA) and γ-aminobutyric (GABA) acids permease. In the present paper, we demonstrate that extracellular amino acids regulate UGA4 expression and that this effect seems to be mediated by the amino acid sensor complex SPS (SSY1, PTR3, SSY5).

Similar content being viewed by others
References
Abdel-Sater F, Iraqui I, Urrestarazu A, André B (2004) The external amino acid signaling pathway promotes activation of Stp1 and Uga35/Dal81 transcription factors for induction of the AGP1 gene in Saccharomyces cerevisiae. Genetics 166:1727–1739
André B, Talibi D, Soussi-Boudekou S, Hein C, Vissers S, Coornaert D (1995) Two mutually exclusive regulatory systems inhibit UASGATA, a cluster of 5′-GAT(A/T)A-3′ upstream from UGA4 gene of Saccharomyces cerevisiae. Nucleic Acids Res 23:558–564
Béchet J, Grenson M, Wiame J-M (1970) Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem 12:31–39
Bermúdez Moretti M, Correa García S, Ramos E, Batlle A (1996) δ-Aminolevulinic acid uptake is mediated by the γ-aminobutyric acid-specific permease Uga4. Cell mol Biol 42:519–523
Bernard F, André B (2001) Genetic analysis of the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. Mol Microbiol 41:489–502
Bricmont PA, Daugherty JR, Cooper TG (1991) The DAL81 gene product is required for induced expression of two differently regulated nitrogen catabolic genes in Saccharomyces cerevisiae. Mol Cell Biol 11:1161–1166
Coffman JA, Rai R, Loprete DM, Cunningham T, Svetlov V, Cooper TG (1997) Cross regulation of four GATA factors that control nitrogen catabolite gene expression in Saccharomyces cerevisiae. J Bacteriol 179:3416–3429
Cooper TG (2002) Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol Rev 26:223–238
Correa García S, Bermúdez Moretti M, Batlle A (2000) Constitutive expression of the UGA4 gene in Saccharomyces cerevisiae depends on two positive-acting proteins, Uga3p and Uga35p. FEMS Microbiol Lett 184:219–224
Cunningham TS, Dorrington RA, Cooper TG (1994) The UGA4 UASNTR site required for GLN3-dependent transcriptional activation also mediates DAL80-responsible regulation and DAL80 protein binding in Saccharomyces cerevisiae. J Bacteriol 178:3470–3479
Didion T, Regenberg B, Jorgensen MU, Kielland-Brandt MC, Andersen HA (1998) The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol Microbiol 27:643–650
Donaton MC, Holsbeeks I, Lagatie O, Van Zeebroeck G, Crauwels M, Windericjx J, Thevelein JM (2003) The gap general amino acid permease acts as an amino acid sensor for activation of protein kinase A targets in the yeast Saccharomyces cerevisiae. Mol Microbiol 50:911–929
Forsberg H, Ljungdahl PO (2001) Genetic and biochemical analysis of the plasma membrane Ssy1p-Ptr3p-Ssy5 sensor of extracellular amino acids. Mol Cell Biol 21:814–826
Forsberg H, Gifstring CF, Zargari A, Martínez P, Ljungdahl PO (2001) The role of the yeast plasma membrane SPS nutrient sensor in the metabolic response to extracellular amino acids. Mol Microbiol 42:215–228
Grenson M, Muyldermans F, Broman K, Vissers S (1987) 4-Aminobutyric acid (GABA) uptake in baker’s yeast Saccharomyces cerevisiae is mediated by the general amino acid permease, the proline permease and a specific permease integrated into the GABA-catabolic pathway. Biochemistry Life Sci Adv 6:35–39
Iraqui I, Vissers S, Bernard F, de Craene J-O, Boles E, Urrestarazu A, André B (1999) Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol Cell Biol 19:989–1001
Jacobs P, Jauniaux J-C, Grenson M (1980) A cis dominant regulatory mutation linked to the argB-argC gene cluster in Saccharomyces cerevisiae. J Mol Biol 139:691–704
Klasson H, Fink GR, Ljungdahl PO (1999) Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol Cell Biol 19:5405–5416
Kodama Y, Omura F, Takahashi K, Shirahige K, Ashikari T (2002) Genome-wide expression analysis of genes affected by amino acid sensor Ssy1p in Saccharomyces cerevisiae. Curr Genet 41:63–72
Kraakman L, Lemaire K, Ma P, Teunissen AW, Donaton MC, Van Dijck P, Winderckx J, de Winde JH, Thevelein JM (1999) A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol Microbiol 32:1002–1012
Kulkarni AA, Abul-Hamd AT, Rai R, El Berry H, Cooper TG (2001) Gln3p nuclear localization and interaction with Ure2 in Saccharomyces cerevisiae. J Biol Cell 276:32136–32144
Lorenz MC, Heitman J (1998) The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J 17:1236–1247
Lorenz MC, Pan X, Harashima T, Cardenas ME, Xue Y, Hirsch JP, Heitman J (2000) The G protein-coupled receptor Gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics 154:609–622
Marini A-M, Soussi-Boudekou S, Vissers S, André B (1997) A family of ammonium transporters in Saccharomyces cerevisiae. Mol Cell Biol 17:4282–4293
Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
Myers AM, Tzagoloff A, Kinney DM, Lusty CJ (1986) Yeast shutlle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene 45:299–310
Özcan S, Dover J, Rosenwald AG, Wolfl S, Johnston M (1996) Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction gene expression. Proc Natl Acad Sci USA 93:12428–12432
Sherman F (1991) Getting started with yeast. Methods Enzymol 194:3–21
Soussi-Boudekou S, Vissers S, Urrestarazu A, Janiaux J-C, André B (1997) Gzf3p, a fourth GATA factor involved in nitrogen regulated transcription in Saccharomyces cerevisiae. Mol Microbiol 23:1157–1168
Acknowledgements
This work was supported by grants from the Argentine National Research Council (CONICET) (PEI6306-2004), the UBACYT X195 (2001–2003) and the UBACYT X014 (2004–2007). Mariana Bermúdez Moretti and Susana Correa García hold the post of Associate Researchers at the CONICET. Alcira Batlle holds the post of Superior Researcher at the CONICET.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bermudez Moretti, M., Perullini, A.M., Batlle, A. et al. Expression of the UGA4 gene encoding the δ-aminolevulinic and γ-aminobutyric acids permease in Saccharomyces cerevisiae is controlled by amino acid-sensing systems. Arch Microbiol 184, 137–140 (2005). https://doi.org/10.1007/s00203-005-0022-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-005-0022-3