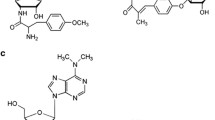
Abstract
The left ends of the biosynthetic gene clusters of novobiocin (nov), clorobiocin (clo) and coumermycin A1 (cou) from Streptomyces spheroides (syn. S. caeruleus) NCIMB 11891, S. roseochromogenes var. oscitans DS 12.976 and S. rishiriensis DSM 40489 were cloned and sequenced. Sequence comparison suggested that novE, cloE and couE, respectively, represent the borders of these three clusters. Inactivation of novE proved that novE does not have an essential catalytic role in novobiocin biosynthesis, but is likely to have a regulatory function. The gene products of novF and cloF show sequence similarity to prephenate dehydrogenase and may produce 4-hydroxyphenylpyruvate (4HPP) as a precursor of the substituted benzoate moiety of novobiocin and clorobiocin. Coumermycin A1 does not contain this benzoate moiety, and correspondingly the coumermycin cluster was found not to contain a functional novF homologue. The coumermycin biosynthetic gene cluster apparently evolved from an ancestral cluster similar to those of novobiocin and clorobiocin, and parts of the ancestral novF homologue have been deleted in this process. No homologue to novC was identified in the gene clusters of clorobiocin and coumermycin, questioning the postulated involvement of novC in aminocoumarin biosynthesis. Heterologous expression of novDEFGHIJK in Streptomyces lividans resulted in the formation of 2,4-dihydroxy-α-oxy-phenylacetic acid, suggesting that at least one of the proteins encoded by these genes may participate in a hydroxylation reaction.



Similar content being viewed by others
Abbreviations
- aac(3)IV :
-
Apramycin resistance gene
- aac-p :
-
Apramycin resistance gene promoter
- oriT :
-
Origin of transfer from RK2
- FRT :
-
FLP recognition target
- 2,4-DHOPA :
-
2,4-Dihydroxy-α-oxy-phenylacetic acid
- 4HPP :
-
4-Hydroxyphenylpyruvate
References
Bigi F, Casiraghi G, Casnati G, Sartori G (1984) Unusual Friedel-Crafts reactions. Part 8. Synthesis of 2-hydroxyarylglyoxylic acids via ortho-specific oxaloylation of phenols with oxalyl chloride. J Chem Soc, Perkin Trans 1:2655–2657
Bunton CA, Kenner GW, Robinson MJT, Webster BR (1963) Experiments related to the biosynthesis of novobiocin and other coumarins. Tetrahedron 19:1001–1010
Calvert RT, Spring MS, Stoker JR (1972) Investigations of the biosynthesis of novobiocin. J Pharm Pharmacol 24:972–978
Chen H, Walsh CT (2001) Coumarin formation in novobiocin biosynthesis: β-hydroxylation of the aminoacyl enzyme tyrosyl-S-NovH by a cytochrome P450 NovI. Chem Biol 8:301–312
Chiu H-T, Hubbard BK, Shah AN, Eide J, Fredenburg RA, Walsh CT, Khosla C (2001) Molecular cloning and sequence analysis of the complestatin biosynthetic gene cluster. Proc Natl Acad Sci U S A 98:8548–8553
Doumith M, Weingarten P, Wehmeier UF, Salah-Bey K, Benhamou B, Capdevila C, Michel J-M, Piepersberg W, Raynal M-C (2000) Analysis of genes involved in 6-deoxyhexose biosynthesis and transfer in Saccharopolyspora erythraea. MGG, Mol Gen Genet 264:477–485
Eustáquio AS, Gust B, Luft T, Li S-M, Chater KF, Heide L (2003) Clorobiocin biosynthesis in Streptomyces : Identification of the halogenase and generation of structural analogs. Chem Biol 10:279–288
Galm U, Schimana J, Fiedler HP, Schmidt J, Li S-M, Heide L (2002) Cloning and analysis of the simocyclinone biosynthetic gene cluster of Streptomyces antibioticus Tü 6040. Arch Microbiol 178:102–114
Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A 100:1541–1546
Hojati Z, Milne C, Harvey B, Gordon L, Borg M, Flett F, Wilkinson B, Sidebottom PJ, Rudd BAM, Hayes MA, Smith CP, Micklefield J (2002) Structure, biosynthetic origin, and engineered biosynthesis of calcium-dependent antibiotics from Streptomyces coelicolor. Chem Biol 9:1175–1187
Holzenkämpfer M, Zeeck A (2002) Biosynthesis of simocyclinone D8 in an 18O2-rich atmosphere. Journal of Antibiotics 55:341–342
Horinouchi S, Beppu T (1994) A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol Microbiol 12:859–864
Hubbard BK, Thomas MG, Walsh CT (2000) Biosynthesis of L-p-hydroxyphenylglycine, a non-proteinogenic amino acid constituent of peptide antibiotics. Chem Biol 7:931–942
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics, 2nd edn. John Innes Foundation, Norwich, UK
Kominek LA (1972) Biosynthesis of novobiocin by Streptomyces niveus. Antimicrob Agents Chemother 1:123-134
Leskiw BK, Bibb MJ, Chater KF (1991) The use of a rare codon specifically during development? Mol Microbiol 5:2861–2867
Maxwell A (1999) DNA gyrase as a drug target. Biochem Soc Trans 27:48–53
Peschke U, Schmidt H, Zhang HZ, Piepersberg W (1995) Molecular characterization of the lincomycin-production gene cluster of Streptomyces lincolnensis 78–11. Mol Microbiol 16:1137–1156
Pojer F, Li S-M, Heide L (2002) Molecular cloning and sequence analysis of the clorobiocin biosynthetic gene cluster: new insights into the biosynthesis of aminocoumarin antibiotics. Microbiology 148:3901–3911
Pojer F, Wemakor E, Kammerer B, Chen H, Walsh CT, Li S-M, Heide L (2003) CloQ, a prenyltransferase involved in clorobiocin biosynthesis. Proc Natl Acad Sci U S A 100:2316–2321
Quiros LM, Aguirrezabalaga I, Olano C, Mendez C, Salas JA (1998) Two glycosyltransferases and a glycosidase are involved in oleandomycin modification during its biosynthesis by Streptomyces antibioticus. Mol Microbiol 28:1177–1185
Retzlaff L, Distler J (1995) The regulator of streptomycin gene expression, StrR, of Streptomyces griseus is a DNA binding activator protein with multiple recognition sites. Mol Microbiol 18:151–162
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York
Schmutz E, Mühlenweg A, Li S-M, Heide L (2003) Resistance genes of aminocoumarin producers: Two type II topoisomerase genes confer resistance against coumermycin A1 and clorobiocin. Antimicrob Agents Chemother 47:869–877
Steffensky M, Li S-M, Vogler B, Heide L (1998) Novobiocin biosynthesis in Streptomyces spheroides: identification of a dimethylallyl diphosphate:4-hydroxyphenylpyruvate dimethylallyl transferase. FEMS Microbiol Lett 161:69–74
Steffensky M, Li S-M, Heide L (2000a) Cloning, overexpression, and purification of novobiocic acid synthetase from Streptomyces spheroides NCIMB 11891. J Biol Chem 275:21754–21760
Steffensky M, Mühlenweg A, Wang Z-X, Li S-M, Heide L (2000b) Identification of the novobiocin biosynthetic gene cluster of Streptomyces spheroides NCIB 11891. Antimicrob Agents Chemother 44:1214–1222
Thamm S, Distler J (1997) Properties of C-terminal truncated derivatives of the activator, StrR, of the streptomycin biosynthesis in Streptomyces griseus. FEMS Microbiol Lett 149:265–272
Trefzer A, Pelzer S, Schimana J, Stöckert S, Bihlmaier C, Fiedler HP, Welzel K, Vente A, Bechthold A (2002) Biosynthetic gene cluster of simocyclinone, a natural multihybrid antibiotic. Antimicrob Agents Chemother 46:1174–1182
van Wageningen AM, Kirkpatrick PN, Williams DH, Harris BR, Kershaw JK, Lennard NJ, Jones M, Jones SJ, Solenberg PJ (1998) Sequencing and analysis of genes involved in the biosynthesis of a vancomycin group antibiotic. Chem Biol 5:155–162
Wang Z-X, Li S-M, Heide L (2000) Identification of the coumermycin A1 biosynthetic gene cluster of Streptomyces rishiriensis DSM 40489. Antimicrob Agents Chemother 44:3040–3048
Acknowledgements
We thank Axel Zeeck for helping on the structural elucidation of 2,4-DHOPA; Helen Kieser, Tobias Kieser, and Sofoklis Lekkas for valuable suggestions for conjugation procedures; and Marion Steffensky for DNA sequencing of the novobiocin cluster. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (to LH and S-ML), and by grant 208/IGF12432 from the Biotechnological and Biological Research Council (to KFC).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Eustáquio, A.S., Luft, T., Wang, ZX. et al. Novobiocin biosynthesis: inactivation of the putative regulatory gene novE and heterologous expression of genes involved in aminocoumarin ring formation. Arch Microbiol 180, 25–32 (2003). https://doi.org/10.1007/s00203-003-0555-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-003-0555-2