Abstract
Background
So far STR (Short Tandem Repeat) profiling of single cells has played hardly any role in forensic molecular-biological casework; however, the application of this technique could open up new possibilities for deducing the unambiguous DNA profile of each single contributor involved in a mixture of two or more components.
Objective
For this purpose DEPArrayTM technology was used to isolate single white blood cells from mixed stains containing blood from two or three different contributors. The STR profiling was carried out on each single cell.
Material and methods
In order to standardize the interpretation of the findings, which, like all low template (LT) DNA analyses, shows characteristic artifacts, leukocytes from a fresh blood sample were examined in advance and the profiles were evaluated. Based on these findings, the results of the examination of three mixed stains (consisting of blood from two or three persons) were evaluated.
Results
In this way, complete or almost complete DNA profiles could be deduced for all persons involved in the corresponding mixture.
Conclusion
The STR profiling of single cells isolated by DEPArrayTM technology opens up completely new possibilities in the field of mixture deconvolution, especially for mixtures composed of cells of the same cell type.
Zusammenfassung
Hintergrund
STR(Short Tandem Repeat)-Analysen von Einzelzellen spielen bislang in der forensisch-molekularbiologischen Fallarbeit kaum eine Rolle. Die Anwendung dieser Technik, im Speziellen zur Ableitung von DNA-Profilen einzelner, an einer Mischung beteiligten Personen, könnte jedoch neue Möglichkeiten eröffnen.
Ziel der Arbeit
Zu diesem Zweck sollen in dieser Arbeit mit Hilfe der DEPArrayTM-Technologie einzelne Leukozyten aus Mischspuren, in denen sich Blut mehrerer Personen findet, separiert einer Einzelzell-STR-Analyse zugeführt werden.
Material und Methode
Um die Interpretation der Befunde zu standardisieren, die wie alle Low-template (LT)-DNA-Analysen charakteristische Artefakte aufweisen, wurden vorab Leukozyten einer frischen Blutprobe untersucht und die Profile ausgewertet. Basierend auf diesen Erkenntnissen wurden im zweiten Schritt die Ergebnisse der Untersuchung von 3 Mischspuren (bestehend aus Blut von 2 bzw. 3 Personen) bewertet.
Ergebnisse
Für alle, an den jeweiligen Spuren beteiligte Personen konnten vollständige bzw. fast vollständige DNA-Profile abgeleitet werden.
Schlussfolgerung
Durch die STR-Typisierung einzelner, mit Hilfe der DEPArrayTM-Technologie, separierter Zellen eröffnen sich vollständig neue Möglichkeiten zur Ableitung von Einzelprofilen aus Mischungen, insbesondere wenn sich die entsprechenden Mischspuren aus Zellen des gleichen Zelltyps zusammensetzen.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mixture deconvolution
Mixture deconvolution is a main topic in forensic DNA investigations, because routine cases involve a lot of mixtures and short tandem repeat (STR) profiling leads to mixed DNA profiles. With the exception of mixed profiles showing a major component or two-person mixtures, from whom one contributor is known and the alleles from the second could be deduced, for most of these mixtures the unambiguous determination of the alleles of a single contributor is not possible. Therefore mixed profiles can mostly be used for direct comparison of profiles from known persons. A comparison with a national database, which is the aim of almost all investigations in cases where no suspect could be determined, is only possible for some selected national databases [4]. In general, for searching national databases, single person profiles are needed or at least preferable. Moreover, mixed profiles are more complicated to interpret and statistically evaluate than single source samples [12, 17]. In summary, deducing single profiles for all contributors of a given mixture would be a great advantage.
For these reasons, several methods were developed by forensic scientists aiming to split mixed stains into their different cellular components. These include the separation of different cell types by laser capture microdissection (LCM) or fluorescence-activated cell sorting (FACS) [6, 18], as well as the time-shift release of the DNA from the different cellular components of a mixture, such as the differential extraction method for sperm-epithelial cell mixtures [10]. For the splitting of mixed stains consisting exclusively of cells of the same cell type, some other methods may be helpful; for example, the use of anti-human AB0 and CD45 antibody-coated microbeads combined with centrifugal separation for the isolation of white blood cells originating from donors with different blood groups [19]. As a further approach, the sex-specific labelling of cells combined with laser microdissection is used for the isolation of cells from a male and a female contributor [1, 2]. Moreover, some probabilistic models included in statistical software programs were used for mixture deconvolution [5, 11].
DEPArrayTM technology
The DEPArrayTM technology (Menarini Silicon Biosystems, Bologna, Italy) has been successfully used for mixture deconvolution by physical isolation of cells using a digital approach. With the help of this technology, single cells, distinguished by immunofluorescent labels and verified by optical imaging, are isolated with the use of a computer-controlled semiconductor dielectrophoretic chip. Cells are inserted into a flow cell and captured in compartments (so-called cages) generated by the activation of hundreds of thousands of microelectrodes distributed on the floor. Once captured and identified, the cells can be independently moved to a collection vial with extreme precision. Thus, pure pools of cells from different cell populations can be separated from a mixture and recovered in separate aliquots. On the other hand, cells belonging to the same cell population can be separated and recovered individually for single cell STR profiling. For forensic applications, the DEPArrayTM forensic sample prep kit (Menarini Silicon Biosystems), which enables the staining of epithelial cells, leucocytes and sperm cells, was specifically developed by Menarini Silicon Biosystems. The proof of principle was demonstrated by Fontana et al. [8]. Moreover, Williamson et al. used the DEPArrayTM technology for the enhanced mixture deconvolution of sexual offence samples [21]. In a former study this technology has been used in the context of chimerism determination after allogenic bone marrow and stem cell transplantation [3]. All DEPArrayTM-related studies published so far were based on the separation of pure pools of cells from different cell populations. This study attempted to connect the separation of individual cells of the same type with single cell STR profiling for the deconvolution of mixtures containing white blood cells from different contributors.
Single cell STR profiling
Single cell STR profiling is not commonly used in forensic DNA investigations, mostly due to the lack of reliable methods for the isolation of single cells from forensic samples. When single cells can be obtained, some literature exists demonstrating the possibility to obtain a profile using different approaches (Geng et al. Single-cell forensic short tandem repeat typing within microfluidic droplets [9]). A more commonly used procedure in order to obtain single source perpetrator DNA profiles from mixed samples, is the collection of individual skin flakes or bio-particles [7, 15]: however single cells, as well as individual skin flakes, contain a very small amount of DNA, which is not sufficient to obtain STR profiles using the routine extraction and amplification protocols. Therefore, special DNA extraction methods (e.g. single tube lysis and amplification) and modified amplification protocols (low template, LT-DNA protocols) were developed [8, 13]. While improving the sensitivity through an enhancement of PCR, LT-DNA protocols often have an influence on the appearance of the profiles; for example, increased stutter peaks and imbalances of the two peaks of a heterozygote loci as well as increased drop-out rates were observed [20]. This in turn entails the development of special guidelines for the interpretation of these profiles.
Study design
In order to obtain single source perpetrator DNA profiles from blood-blood mixtures of two or three contributors, for this study individual white blood cells (WBC) were separated using the DEPArrayTM technology. The DNA of each single cell was set free in solution with the help of a single tube lysis kit (DEPArrayTM LysePrep Kit, Menarini Silicon Biosystems) and STR profiling was carried out just adding the PCR mix to the lysed cell solution. To get a first impression regarding the quality and reproducibility of single cell profiles in optimal conditions, white blood cells from a fresh blood sample were processed in advance (sample I). Following this first control sample, three additional bloodstains (including mock and real casework samples) were investigated, which contained blood from two or three contributors (samples II–IV). To check the correctness of the obtained single source perpetrator DNA profiles and therefore to demonstrate the proof of principle, only mixtures from known contributors were investigated.
Material and methods
Material
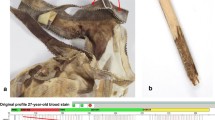
Sample I was a fresh EDTA blood sample, from which 5 µl were removed for investigation. Sample II was a blood stain on the blade of a knife (Fig. 1). The knife was submitted in the context of the investigation in a current murder case. The age of the stain was approximately 6 weeks. Routine STR profiling carried out on several samples taken from the blade as well as the handle of the knife revealed completely matching mixtures. These mixtures could be attributed to two persons and completely explained by the alleles from the victim and suspect. The corresponding reference samples were available. For DEPArrayTM separation another sample was taken from the blade with a nylon flocked, PBS buffer-soaked swab. Sample III was a blood-blood mixture of two individuals (without a clearly recognizable major component) applied on a piece of cellulose, which was part of the German DNA Profiling (GEDNAP) proficiency test of the year 2017 (GEDNAP 55, stain 2). For sample IV a mixture was made up of 5 µl fresh EDTA blood from three different individuals. From this mixture 5 µl were taken and added to the staining procedure. All fresh EDTA blood samples used here were submitted as reference samples for routine cases and therefore the corresponding profiles were known. In order to check the correctness, the obtained single source perpetrator DNA profiles for the GEDNAP proficiency test stain (sample III) were sent to the organizer for confirmation.
Cell separation using DEPArrayTM technology
Cells were resuspended from the cellulose (sample III) or the swab (sample II) according to the manufacturer’s instructions. The resulting cell suspensions as well as the 5 µl whole blood samples (sample I and sample IV) were added to the staining procedure. White blood cells were stained using the Forensic Sample Prep Kit (Menarini Silicon Biosystems) according to the manufacturer’s instructions. For the staining of white blood cells, this kit contains a PE (Phycoerythrin)-conjugated anti-human CD45 antibody. Furthermore, the nuclei were stained with DAPI (4’,6-Diamidin-2-phenylindol). The samples were loaded into DEPArray cartridges and processed according to the manufacturer’s specifications for the DEPArray TM V2 system. White blood cells were classified with the criteria “InCage” (position of the cell inside the dielectrophoretic cage so that it can be moved by the system), DAPI positive and PE positive using the CellBrowser software (Menarini Silicon Biosystems). The maximum number of cells that can be recovered per experiment is determined by the type of chip used and by the type of recovery (single or pooled). On the DEPArray TM V2 system, a maximum of 17 single cells can be isolated, therefore, for samples II, III and IV, 17 single cells were recovered. For sample I a pool of 5 WBCs (as a control sample, which contains five times the amount of DNA and therefore sufficient DNA to yield a full STR profile) as well as 11 single WBCs were isolated.
DNA extraction and STR profiling
The DNA was isolated with the DEPArrayTM LysePrep Kit (Menarini Silicon Biosystems) according to manufacturer’s instructions. Using the Multiplex-PCR PowerPlex® ESXfast system (Promega, Madison, WI, USA) the sex-determining amelogenin system as well 16 autosomal loci were amplified on a Veriti Thermal Cycler (Thermo Fisher Scientific, Waltham, MA, USA). The PCR was carried out in a reaction volume of 14 µl and a 32 cycle PCR program, according to the in-house validated protocol; apart from that, the manufacturer’s instructions were followed. Determination of fragment length was performed on a 3500xl Genetic Analyzer (Thermo Fisher Scientific) according to manufacturer’s instructions. Data analysis was carried out using the GeneMapper® ID-X Software v1.4 (Thermo Fisher Scientific) and a detection threshold of 50rfu.
Results and discussion
Sample I
For all 11 WBCs, profile completeness varied from 18 to 31 of the expected 32 alleles (Table 1). This means that the worst profile still showed more than 50% of the expected alleles, whereas on average approximately 82% of the alleles could be obtained per single cell. While a complete profile could not be obtained for any of the 11 single cells, it can be stated that each individual allele was detectable at least 6 times and 3 alleles were shown in each of the 11 profiles. For the pool of five WBCs a balanced and full profile was obtained.
Fig. 2 shows as an example of the partial profile from one single cell (SC-6 WBC) with one allelic drop-out in locus D16S539 (indicated with*). Beside this drop-out some other characteristics, typically generated by LT protocols, could be observed. For example, some of the alleles of the heterozygote loci show strong imbalances (indicated with arrows). The peak height of the smaller allele is approximately 50% lower or more (50% allele 31 in locus D21S11, 81% allele 13 in locus D10S1248 and 64% allele 17 in locus VWA) in comparison with the second allele of the same locus. The maximum imbalance obtained for the 2 alleles of a heterozygote locus in all 11 profiles showed a ratio of 93.4:6.6% (data not shown). For the interpretation of all subsequent profiles, smaller alleles of a heterozygote locus, which did not reach a peak height of at least 50% of the second allele of the same locus, were not classified as true alleles.
Increased n-4/n-3 stutter was often observed, as well. For example, in the profile for SC-6 WBC (Fig. 2, circled) minus one repeat stutters greater than 15% could be found (stutter to allele 17 in locus D3S1358 15.1%, stutter to allele 17 in locus VWA 29.3% and stutter to both alleles of D12S391 with 16.2 and 20.7%, respectively). The highest peak that occurred in n-4 stutter position showed a peak height ratio of 78.9% (SC-8 WBC) and therefore was classified as allelic drop-in in advance. This peak was not included in further considerations regarding the expression of stutter peaks. The maximum stutter values observed at all other alleles were determined per locus and are listed in Table 2. As a comparison, the maximum stutter values reported by McLaren et al. in their PowerPlex® ESXfast validation study are shown [14]. McLaren et al. calculated the stutter values (mean, standard deviation and maximum stutter ratio) for each locus under consideration of the profiles from 656 individuals. All these profiles were generated using sufficient amounts of DNA and a 30-cycle PCR program. In contrast, the number of profiles created in this study as a part of the feasibility study is still too small to provide reliable results for a detailed statistical analysis. For this reason, the calculations on the determination of the maximum values per locus were initially limited, which should serve as a first idea of the interpretation of the profiles from the following samples. For routine use of this technique and standardized interpretation of the resulting profiles, a special set of appropriate guidelines, based on a bigger data set and a more detailed analysis, has to be developed. McLaren et al. obtained maximum stutter values up to 19.3%, while, in this single cell data set, extremely high stutter up to 34% was detected; for 7 of the 16 autosomal loci investigated individual stutter peaks values higher than 20% were reached. Also, two additional alleles in plus-one repeat stutter position were observed (allele 19 in D18S11, peak height ratio 16.4% and allele 12 in D16S539 with a ratio of 31.1%). Based on this knowledge, for the interpretation of all profiles obtained for samples II, III and IV, peaks in plus-one and minus-one stutter position were only called as true alleles when the peak height ratio was above 35%. Further additional alleles, which were not located in a minus or plus one repeat stutter position did not occur.
Under consideration of possibly occurring increased stutter peaks and the fact that each real individual allele was detectable at least 6 times in the 11 partial single cell profiles (while drop-in alleles were only detected as unique events), a clear and full profile can be deduced by the combined analysis of all the 11 single cells.
Sample II
The profiles of 4 of the recovered 17 white blood cells showed no alleles at all. For the remaining 13 cells partial STR profiles were obtained. These 13 partial profiles could be assigned to the 2 reference profiles, 6 to the victim and 7 to the suspect profile (Table 3). The obtained partial profiles showed 11 up to 27 of the expected 29 (victim) and 14 up to 28 of the expected 30 (suspect) alleles. Combining the alleles assigned to each person in each single cell profile, the full profiles of two individuals could be deduced. Every single allele was detected in at least three out of six (profiles assigned to the victim) or seven (profiles assigned to the suspect) cells. The highest number of allelic drop-out could be observed in the STR-loci VWA, TH01, D8S1179 and D21S11 and thus the observed allelic drop-outs are more likely to be a consequence of stochastic phenomena [16] rather than the consequence of DNA degradation. Moreover, three additional alleles (classified as allelic drop-ins) were observed (Table 3), one of which (allele 29 in locus D21S11, sample SC-5 WBC) was located in a minus one stutter position of the actual suspects’ allele 30. Nevertheless, due to the peak height ratio of approximately 45% this peak was also rated as a drop-in. All three drop-in alleles occurred only once in three different samples and loci, where two other alleles had already been proved several times and therefore the true genotype of this individual could be clearly reconstructed. In summary, complete profiles could be derived for both persons involved in the mixture.
Sample III
In contrast, from the 17 WBCs recovered from the GEDNAP stain (blood-blood mixture from two individuals) only two partial profiles could be obtained (Figs. 3 and 4). The profiles showed different alleles and based on the assumption, that no allelic drop out occurred in the amelogenin, these are the profiles of a male (Fig. 3) and a female (Fig. 4) person. For the remaining cells, no alleles could be detected at all. Under use of the above established values for stutter interpretation, clear signals were labelled with the corresponding allele naming. For the male profile the two alleles in locus D10S1248 and D2S1338 showed extreme imbalances (both not in plus or minus one repeat stutter position). Therefore, the smaller alleles were classified as questionable and not as true alleles. The size of allele 6, (locus TH01) and allele 18 (locus D12S391) in the female profile, as well as allele 14 (locus D19433) in the male profile show peak heights which were about twice as high as the peaks of the neighboring heterozygote loci. For these loci homozygote genotypes were reported. In this way two profiles were deduced; one male profile with at least two questionable and four possibly missing alleles (drop-outs) and a female profile with at least four possibly missing alleles (drop-outs). The deduced partial genotypes were completely confirmed by the organizer of the GEDNAP proficiency test. In summary, although the results are based on single analyses, the derived, albeit partial profiles could be correctly deduced.
Male profile of white blood cells (WBC) SC-17 from sample III (GEDNAP 55, stain 2) evaluated under use of the established values for the classification of true alleles (in stutter position >35%, in cases of heterozygote imbalances smaller peak >50%). The deduced genotype for all loci can be found in the corresponding text boxes. Possible drop-out positions (where a distinction between the presence of an allelic drop-out or an actual homozygote genotype was not possible) are indicated with *
Female profile of white blood cells (WBC) SC-17 from sample III (GEDNAP55, stain 2) evaluated using the established values for the classification of true alleles (in stutter position >35%, in cases of heterozygote imbalances smaller peak >50%). The deduced genotype for all loci can be found in the corresponding text boxes. Possible drop-out positions (where a distinction between the presence of an allelic drop or an actual homozygote genotype was not possible) are indicated with *
In comparison to sample I and II it is obvious that only 2 of the 17 cells could be profiled. This fact did not meet the expectations after the first two attempts. Looking for an explanation, the images of the selected cells from sample I and III were compared (Fig. 5). Whereas the images of the WBCs from sample I showed clear, well-demarcated images for the PE as well as bright field and DAPI staining, the signals of most of the WBCs from sample III appeared more diffuse. This may give a hint on the constitution of the cells and therefore maybe also the DNA quality. According to the information provided by the organizer of the GEDNAP proficiency test, at the time of investigation the stain was approximately 2 years old and therefore older than samples I and II. In addition, it was communicated that the mixed samples of the proficiency test were mostly treated with surfactants. This also may have an influence of the integrity of the cell or nuclear membrane. Damage of the cell membrane in turn may have an influence on the staining and the perfect movement of the cells in the extraction cartridge. Also, a loss of DNA would be conceivable. Whether there really is a connection has to be resolved by further testing.
Sample IV
For all of the recovered 17 WBCs from sample IV, partial profiles were obtained (Table 4) and 3, 6 and 8 profiles can be assigned to an individual (named A, B and C, respectively). The obtained partial profiles showed 21 up to 29 of the expected 31 (A), 24 up to 29 of the expected 33 (B) and 18 up to 22 of the expected 23 (C) alleles. A total of 16 alleles were detected only once and therefore not classified as true alleles (possible drop-in alleles). All other alleles were detected at least three out of six (A), two out of three (B) or five out of 8 (C) times. Of the 16 alleles proven only once 13 occurred in loci where 1 (in the case of a homozygote genotype) or 2 (in the case of a heterozygote genotype) other alleles had already been proven several times. Therefore, the true genotype of this individual could be deduced. The other three alleles were found in the three profiles assigned to individual B (allele 18 in locus VWA and alleles 14.2 and 15 in locus D19S433). A comparison with the reference profile of the corresponding person showed that these were indeed the true alleles of the corresponding individuals. So once more it was shown that for the certain and complete derivation of a genotype, a sufficiently large number of partial profiles should be used in order to distinguish artifacts from true alleles. Thereby the number of the required profiles directly correlates with the quality.
For this sample, two complete and one almost complete partial profile were finally deduced, which all completely matched the alleles of the reference samples and therefore demonstrated the effectiveness and correctness of this method. Sample IV again was a fresh and untreated sample. Compared to sample III, significantly better typing results (partial profiles for all 17 recovered WBCs) could be obtained. This again underlines the presumption that the success of the technology seems to depend very much on the quality of the cells and/or DNA.
Conclusion
In summary, it can be stated that STR profiling of single cells isolated by DEPArrayTM technology can be used successfully for the deconvolution of mixtures containing white blood cells from different contributors. While the benefits of the technology in the separation of heterogeneous mixtures composed of different cell types have already been demonstrated impressively several times [3, 8, 21], in the current study for the first time DNA profiles of each individual contributor could be reconstructed from mixtures composed of cells of the same type through the isolation and profiling of multiple single cells.
In contrast to software-based deconvolution models, this approach is well-suited for the investigation of mixtures with balanced mixture ratios. Software-based models perform deconvolution by utilizing the peak height information for inferring the most likely profile genotypes for the unknown contributors [5]. This can be used very well for deducing the alleles of a major component; however, the more balanced a mixture is the less meaningful are the calculated probabilities for the possible genotypes of the different contributors. In addition, for samples with very low amounts of DNA, reliable estimations could not be achieved because the peak height levels are too much influenced by stochastic variations. Therefore, depending on the mixture ratio and amount of template DNA, STR profiling of single cells isolated by DEPArrayTM may be an advantage.
From the results of sample III it can be concluded that the success of the technology presented here, however, seems to depend very much on the quality of the cells and/or DNA. Single cell STR profiling itself is already a challenge, even when DNA is intact. Degradation or loss of DNA may very quickly lead to completely negative results. Furthermore, the structure of the cell membrane must be preserved to a level that the staining and the movement of the cells in the cartridge is still possible without restriction.
The single cells treated in this study yielded only partial profiles. In order to obtain complete profiles, all partial profiles (if available), which were attributed to the same single person were combined to form a consensus sequence. Artifacts can also be better recognized in this way, so that the true alleles can be deduced beyond doubt. Therefore, with increasing complexity of the mixture (increasing number of contributors), the number of selected cells has to be increased. With the DEPArrayTM V2 system used in this study, a maximum of 17 single cells can be recovered from 1 cartridge. In contrast, the new generation, DEPArrayTMN xT instrument, allows separation of up to 48 single cells.
As expected, when low amounts of DNA were amplified using an increased PCR cycle program, beside drop-outs some other characteristic artefacts occurred more often. Initial guideline values were determined in the course of this feasibility study; however, for the routine application of this technology in forensic casework, appropriate guidelines should be developed based on a larger dataset. With the help of such guidelines, STR profiling of single cells isolated by DEPArrayTM technology opens up completely new possibilities in the field of mixture deconvolution.
References
Anslinger K, Mack B, Bayer B, Rolf B, Eisenmenger W (2005) Digoxigenin labelling and laser capture microdissection of male cells. Int J Legal Med 119:374–377
Anslinger K, Mack B, Bayer B, Eisenmenger W (2007) Sex-specific fluorescent labelling of cells for laser microdissection and DNA profiling. Int J Legal Med 121:54–56
Anslinger K, Bayer B, von Máriássy D (2018) Application of DEPArrayTM technology for the isolation of white blood cells from cell mixtures in chimerism analysis. Rechtsmedizin 28:134–137
Benschop CCG, van de Merwe L, de Jong J, Vanvooren V, Kempenaers M, Kees van der Beek CP, Barni F, Reyes EL, Moulin L, Pene L, Haned H, Sijen T (2017) Validation of SmartRank: a likelihood ratio software for searching national DNA databases with complex DNA profiles. Forensic Sci Int Genet 29:145–153
Bleka Ø, Benschop CCG, Storvik G, Gill P (2016) A comparative study of qualitative and quantitative models used to interpret complex STR DNA profiles. Forensic Sci Int Genet 25:85–96
Elliott K, Hill D, Lambert C, Burroughes T, Gill P (2003) Use of laser microdissection greatly improves the recovery of DNA from sperm on microscope slides. Forensic Sci Int 137:28–36
Farash K, Hanson EK, Ballantyne J (1997) Enhanced genetic analysis of single human bioparticles recovered by simplified micromanipulation from forensic “touch DNA”evidence. J Vis Exp 9. https://doi.org/10.3791/52612
Fontana F, Rapone C, Bregola G, Aversa R, de Meo A, Signorini G, Sergio M, Ferrarini A, Lanzellotto R, Medoro G, Giorgini G, Manaresi N, Berti A (2017) Isolation and genetic analysis of pure cells from forensic biological mixtures: the precision of a digital approach. Forensic Sci Int Genet 29:225–241
Geng T, Novak R, Mathies RA (2014) Single-cell forensic short tandem repeat typing within microfluidic droplets. Anal Chem 86:703–712
Gill P, Jeffreys A, Werrett D (1985) Forensic application of DNA “fingerprints”. Nature 318:577–579
Gill P, Sparkes R, Pinchin R, Clayton T, Whitaker J, Buckleton J (1998) Interpreting simple STR mixtures using allele peak areas. Forensic Sci Int 9:41–53
Gill P, Brenner CH, Buckleton JS, Carracedo A, Krawczak M, Mayr WR, Morling N, Prinz M, Schneider PM, Weir BS (2003) DNA commission of the International Society of Forensic Genetics: recommendations on the interpretation of mixtures; DNA commission of the international society of forensic genetics. Forensic Sci Int 160:90–101
Marshall PL, King JL, Budowle B (2015) Utility of amplification enhancers in low copy number DNA analysis. Int J Legal Med 129:43–52
McLaren RS, Bourdeau-Heller J, Patel J, Thompson JM, Pagram J, Loake T, Beesley D, Pirttimaa M, Hill CR, Duewer DL, Kline MC, Butler JM, Storts DR (2014) Developmental validation of the powerPlex(®) ESI 16/17 fast and powerPlex(®) ESX 16/17 fast systems. Forensic Sci Int Genet 13:195–205
Schneider H, Sommerer T, Rand S, Wiegand P (2011) Hot flakes in cold cases. Int J Legal Med 125:543–548
Timken MD, Klein SB, Buoncristiani MR (2014) Stochastic sampling effects in STR typing: implications for analysis and interpretation. Forensic Sci Int Genet 11:195–204
Ulbrich W, Anslinger K, Bäßler G, Eckert M, Fimmers R, Hohoff C, Kraft M, Leuker C, Molsberger G, Pich U, Razbin S, Schneider H, Templin M, Wächter A, Weirich V, Zierdt H, Schneider PM (2016) Gemeinsame Empfehlungen der Projektgruppe „Biostatistische DNA-Berechnungen“ und der Spurenkommission zur biostatistischen Bewertung von DNA-analytischen Befunden. Rechtsmedizin 26:291–298
Verdon TJ, Mitchell RJ, Chen W, Xiao K, Van Oorschot RAH (2015) FACS separation of non-compromised forensically relevant biological mixtures. Forensic Sci Int Genet 14:194–200
Yano S, Honda K, Kaminiwa J, Nishi T, Iwabuchi Y, Sugano Y, Kurosu A, Suzuki Y (2014) DNA extraction for short tandem repeat typing from mixed samples using anti-human leukocyte CD45 and ABO blood group antibodies. Forensic Sci Int Genet 10:17–22
Whitaker JP, Cotton EA, Gill P (2001) A comparison of the characteristics of ptofiles produced with the AMPFISTR SGM Plus multiplex system for both standard and low copy number (LCN) STR DNA analysis. Forensic Sci Int 123:215–223
Williamson VR, Laris TM, Romano R, Marciano MA (2018) Enhanced DNA mixture deconvolution of sexual offense samples using the DEPArray™ system. Forensic Sci Int Genet 34:265–276
Acknowledgements
The authors thank Menarini Silicon Biosystems for the provision of the DEPArrayTM system as well as for the valuable support and Eppendorf for providing necessary laboratory equipment.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
K. Anslinger, M. Graw and B. Bayer declare that they have no competing interests.
The investigations were carried out according to the specifications of the Central Ethics Committee of the Federal Medical Council.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Anslinger, K., Graw, M. & Bayer, B. Deconvolution of blood-blood mixtures using DEPArrayTM separated single cell STR profiling. Rechtsmedizin 29, 30–40 (2019). https://doi.org/10.1007/s00194-018-0291-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00194-018-0291-1