Abstract
Objective
To evaluate the accuracy of the soluble triggering receptor expressed on myeloid cells-1 (sTREM-1) as a diagnostic test for bacterial infection.
Design
Meta-analysis of 13 diagnostic studies.
Data source
Medline; Embase; Web of Science (from January 1966 to January, update to August 2008); and Cochrane Controlled Clinical Trials Register Database (through first quarter 2008).
Measurements and results
A meta-analysis of all 73 studies was performed. Thirteen studies fulfilled the inclusion criteria (980 patients, 557 patients with bacterial infection, 423 with non-bacterial infection); global prevalence was 56.8%. The global sensitivity was 0.82 (95% confidence interval CI, 0.68–0.90), the specificity was 0.86 (95% CI, 0.77–0.91), the positive likelihood ratio (PLR) was 5.66 (95% CI, 3.41–9.38), the negative likelihood ratio (NLR) was 0.21 (95% CI, 0.12–0.40), and the diagnostic odds ratio (DOR) was 26.35 (95% CI, 10.32–67.28). The area under the curve of the summary receiver operator characteristic (SROC) was 0.86 (95% CI, 0.77–0.91), with a Q point value of 0.84. The sensitivity of the sTREM-1 assay for diagnosis of urinary tract infection was low (0.18, 95% CI, 0.05–0.51).
Conclusions
sTREM-1 represents a reliable biological marker of bacterial infection, but it may be not a sufficient biological marker for infection of the urinary tract as a result of its low sensitivity. Whether sTREM-1 guidance can reduce antibiotic use as well as the measurement of sTREM-1 in different types of infection will require additional prospective studies.
Similar content being viewed by others
Introduction
Bacterial infections and sepsis are a common cause of morbidity and mortality in intensive care units (ICUs) [1, 2]. The diagnosis of sepsis is difficult because clinical signs of sepsis often overlap with other non-infectious causes of systemic inflammation [3, 4]. In addition, diagnosis of bacterial infections is sometimes challenging because clinical presentation of infections from different causative agents can be similar. The ability to accurately and rapidly identify patients with bacterial infections is an important management issue for ICU physicians. Accurate and timely diagnosis could guide treatments, reduce misuse of antibiotics, limit morbidity, and possibly improve patients’ long-term outcomes [5, 6]. Microbiological cultures can be used to distinguish sepsis from non-infectious conditions. However, this method lacks sensitivity and specificity; and there is often a substantial time delay due to the previous use of antibiotics [7].
There have been attempts to use biological markers in the diagnosis of bacterial infections [2, 6, 8]. Among the potentially useful markers of infection, procalcitonin has been suggested to be the most promising. However, recent meta-analyses show that procalcitonin has a low diagnostic performance in differentiating sepsis from SIRS in critically ill adult patients or moderate diagnostic ability for bacterial infections [9–11]. Serum C-reactive protein is a widely used technique that is low in cost but influenced at early timepoints by other inflammatory circumstances [7, 11].
The triggering receptor expressed on myeloid cells-1 (TREM-1) is a recently discovered member of the immunoglobulin super family, the expression of which on phagocytosis is up-regulated by exposure to bacteria and fungi [12]. Currently, the ligand for TREM-1 is unknown, but through the use of an agonist antibody it was found that TREM-1 signaling induces the production of pro-inflammatory cytokines and chemokines, reactive oxygen species (ROS), and leads to rapid degranulation of neutrophilic granules, and phagocytosis [13]. TREM-1 can be found in its soluble form (soluble triggering receptor expressed on myeloid cells-1, sTREM-1) in body fluids [12–15]. Although promising, the clinical use of sTREM-1 and whether sTREM-1 could be useful in different infectious conditions requires more evidence.
We performed the present meta-analysis to assess the diagnostic use of sTREM-1 and to establish the overall accuracy of sTREM-1 measurement for the diagnosis of bacterial infection.
Materials and methods
Search strategy and study selection
We searched the following electronic databases: Medline; Embase; Web of Science (from January 1966 to January, 2008); and Cochrane Controlled Clinical Trials Register Database (through first quarter 2008) for all studies examining the diagnostic accuracy of sTREM-1 for bacterial infection. We searched electronic databases update to August 2008. For the electronic search, we used the following terms or MeSH subject headings: “TREM1 protein, human”, “triggering receptor expressed on myeloid cells-1”, “soluble triggering receptor expressed on myeloid cells-1”, “TREM-1”, “sTREM-1”; “bacteremia”, “septicemia”, “bacterial infection”, “microbial infection”, “sepsis”, “septic shock”, “sepsis syndrome”, “infection” “systemic inflammatory response syndrome”, and “SIRS”; and “diagnosis”, “sensitivity”, “specificity”, “predictive value”, “likelihood ratio”, “review”, “meta-analysis”, “false positive”, and “false negative”.
We contacted the authors for further study details if needed and searched the reference lists from primary and review articles. No language restriction was used, and all foreign language publications were translated. Further searches were performed by manually reviewing abstract booklets, conference proceedings, and review articles.
We included all studies that met the following criteria: assessing the diagnostic accuracy of sTREM-1 for bacterial infection; providing both sensitivity (true-positive rate) and specificity (true-negative rate) of sTREM-1 for the diagnosis of bacterial infection; providing sufficient information to construct the 2 × 2 contingency table for individual study subjects; and stating a correct test for sTREM-1 in the methods, which was determined in the studies (e.g., enzyme-linked immunosorbent assay, immunoblot technique). We excluded conference abstracts, abstracts and letters because of limited information. Two reviewers (Jing JY and Huang TC) independently judged study eligibility while screening the citations. Disagreements were resolved by consensus.
Data extraction
The final set of English language articles was assessed independently by two reviewers (Jing JY and Huang TC). The reviewers independently abstracted data in each study to obtain information on the year of publication, country of origin, clinical setting, patients’ demographics, sample size, test methods, diagnostic cut-off points, participant characteristics, sensitivity and specificity of the data, and methodological quality. Each reviewer extracted the data to construct a 2 × 2 table. Any disagreements were resolved by consensus.
Quality assessment
The methodological quality of each study was assessed using a checklist based on criteria adapted from the Cochrane Collaboration guidelines [16] and the quality assessment for studies of diagnostic accuracy (QUADAS) tool (maximum score 14) [17].
Studies were grouped according to Sackett and Haynes’ [18] classification of diagnostic studies. In this classification, phase 1 studies are those that compare the difference in test results between patients with the target disorder and healthy individuals. Phase 2 studies are those that examine how the index test discriminates between patients with and without the target disorder. Phase 3 studies are those that assess the test’s real-life performance in patients suspected of having the disorder.
Statistical analysis
We used standard methods recommended for meta-analyses of diagnostic test evaluations [16]. For each study, the sensitivity, specificity, positive and negative likelihood ratios, and a diagnostic odds ratio (OR) were calculated. The diagnostic OR is the ratio of the odds of a positive result in a patient with bacterial infection compared with a patient without bacterial infection: [sensitivity/(1 − sensitivity)]/[(1 − specificity)/specificity]. Each study was weighted using an inverse variance method. We constructed summary receiver operator characteristic (SROC) curves to summarize the study results. A smoothed curve was then fitted across the studies to represent the relationship between sensitivity and the proportion of false positives (1 − specificity). The sensitivity and specificity for the single test threshold identified for each study were used to plot an SROC curve. Pooling of the summary indices was performed using DerSimonian and Laird’s [19] random-effects model.
To detect heterogeneity, the likelihood ratios and diagnostic ORs were graphically displayed using forest plots and analyzed using Cochran’s Q test. A P value of less than 0.05 by Cochran’s Q test indicated significant heterogeneity. To quantify the extent of heterogeneity, the I² statistic was used to measure the percentage of variability among summary indices that were caused by heterogeneity rather than chance. A study with an I² greater than 50% indicated substantial heterogeneity.
To explore sources of heterogeneity among studies, univariate meta-regression analysis (inverse variance weighted) was used. The covariates included spectrum characteristics (e.g., study setting, prevalence, type of bacterial infection), quality of the study (QUADAS scores), and methodological features (e.g., sample size).
Publication bias was examined visually by inspecting funnel plots and statistically by using Egger’s regression model [20]. If publication bias was present, the effect of such a bias on the final summary estimate was assessed using the trim and fill method [21]. This method imputes the missing studies and re-calculates a new summary estimate. The difference between the calculated and observed values was then used to determine the effect of bias on the diagnostic performance of the test. Analyses were performed using Stata (version 10.0; Stata Corporation) and Meta-Test (version 0.6; By Joseph Lau).
Results
Study characteristics
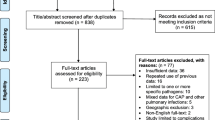
We retrieved 73 abstracts, and 19 publications dealing with sTREM-1 for the diagnosis of bacterial infection were considered as potentially suitable for inclusion in the analysis. After full-text review, seven studies were excluded (Fig. 1): three studies [22–24] were excluded because the sTREM-1 concentration was determined only in the patients with infection, and three studies [25–27] were excluded because the researchers could not generate 2 × 2 tables, one was excluded because it detected the TREM-1 mRNA not sTREM-1 [28]. One was included updated to August 2008 [29]. Totally, 13 studies [29–41] including 557 patients with bacterial infections and 423 non-bacterial infection patients were available for the final analysis. Studies were grouped according to Sackett and Haynes’ classification for diagnostic studies: six Phase 2 studies (group 1), five Phase 3 studies (group 2), and two mixed phase studies (Phases 2 and 1) (group 3).
A total of 980 patients were included in the analysis, comprising 7 studies with 671 patients from intensive care units, one study with 151 patients from emergency departments, and four studies with 158 patients from hospital wards. In five studies (38.4%), the samples were collected from consecutive patients, and, in another four studies (30.8%), the samples were collected with prospective recruitment. Bacterial infections included pneumonia, para-pneumonic effusion, empyema, sepsis, bacterial meningitis, and urinary tract infection. The median age of the patients in the studies was 60 years (interquartile range 46–67 years). The prevalence of bacterial infections across the studies ranged from 30 to 84%. Twelve studies used the enzyme-linked immunosorbent assay (ELISA), and one study used the immunoblot technique. The test threshold ranged from 5 to 374 pg/mL. The details of all 13 studies are shown in Table 1.
Quantitative data synthesis
Figure 2 shows the forest plot for the sensitivity and specificity of 13 sTREM-1 assays used for the diagnosis of bacterial infection. The sensitivity ranged from 0.66 to 0.98 (mean 0.82; 95% CI, 0.68–0.90), while the specificity ranged from 0.40 to 1.00 (mean 0.86; 95% CI, 0.77–0.91). We also noted that the positive likelihood ratio (PLR) was 5.66 (95% CI, 3.41–9.38), the negative likelihood ratio (NLR) was 0.21 (95% CI, 0.12–0.40; Fig. 3), and the DOR was 26.35 (95% CI, 10.32–67.28). Cochran’s Q and I² for sensitivity, specificity, PLR, NLR, and DOR were 227.38 (P < 0.001), 86.93 (P < 0.001), 122.66 (P < 0.001), 339.40 (P < 0.001), and 59.40 (P < 0.001), respectively, and I² for sensitivity, specificity, PLR, NLR, and DOR was 94.72, 86.20, 86.12, 96.49, and 96.63, respectively, indicating significant heterogeneity between studies.
Unlike the traditional ROC plot that explores the effect of varying thresholds (i.e., cut points for determining positive tests) on sensitivity and specificity in a single study, each data point in the SROC plot represents a separate study. The SROC curve presents a global summary of test performance, and it shows the tradeoff between sensitivity and specificity. A graph of the SROC curve for sTREM-1 showing true-positive rates versus false-positive rates from individual studies is shown in Fig. 4. The SROC curve (Fig. 4) yielded a maximum joint sensitivity and specificity of 0.86 (95% CI, 0.77–0.91), an area under the curve of 0.91 (95% CI, 0.88–0.93), and a Q point of 0.84, indicating a median level of overall accuracy.
Multiple regression analysis and publication bias
Spectrum characteristics, the quality of the study, and methodological features were used in the meta-regression analysis to assess the source of variability among studies. As shown in Tables 2 and 3, higher quality studies (QUADAS score, 10) produced sensitivity and specificity values that were not significantly higher than lower quality studies. There were significant differences between classified groups of the diagnostic study, indicating that the classification of diagnostic study affects the diagnostic accuracy. There were significant differences between different study settings of the diagnostic study in diagnostic specificity, indicating that the study setting also potentially affects the diagnostic specificity. Study designs, sample size, and prevalence did not affect the diagnostic accuracy, except for the observation that the consecutive recruitment potentially affected the diagnostic sensitivity (0.66 95% CI, 0.40–0.84; P = 0.05). As to the types of bacterial infections, the sensitivity of the sTREM-1 assay for diagnosis of urinary tract infection was low (0.18, 95% CI, 0.05–0.51; P < 0.001).
Publication bias was detected using Egger’s regression mode. The result of the Egger’s test was not significant (P = 0.059). Visual inspection of the funnel plot for publication bias also suggested that the studies were symmetry on the summary estimate (Fig. 5). These results indicated no publication bias.
Discussion
The present meta-analysis complied with the recommendations for reporting meta-analyses of diagnostic tests [42, 43]. All studies included in the analysis used samples obtained from the location of the infection. Like other similar cell-surface receptors, TREM-1 has a short intracellular domain and, when bound to its as-yet-unidentified ligand, associates with a signal-transduction molecule, DAP12, and triggers the secretion of inflammatory cytokines that amplify the host response to microbial agents [12, 13, 15]. Microbial ligands such as lipopolysaccharide (LPS) increase TREM-1 expression via a post-transcriptional mechanism [44]. TREM-1 mediates the acute inflammatory response to microbial products. Human tissues infected with bacteria are infiltrated with neutrophils and monocytes that express high levels of TREM-1. Conversely, TREM-1 is only weakly expressed in samples from patients with non-infectious inflammatory disorders. TREM-1 is also shed by the membranes of activated phagocytes and can be found in its soluble form in body fluids [12–15]. Excluding one study [28], most of the current studies detected the soluble form of TREM-1 (sTREM-1), and the sTREM-1 assays were performed using sample material from suspected sites of infection.
The results of this systematic review and meta-analysis indicate that the sTREM-1 test can be used as a helpful diagnostic criterion for bacterial infection. Using pooled data from 13 studies comprising 980 subjects, we found the area under the summary receiver operating characteristic to be 0.91, with a lower limit 95% CI of 0.88, indicating a high level of predictive ability for sTREM-1 in the diagnosis of bacterial infection. With a pretest probability of 40%, using a sTREM-1 test would raise the post-test probability to 79%. With a negative likelihood ratio of 0.21, application of a sTREM-1 test would reduce the post-test probability to 0.13, which is helpful for ruling out a possible infection (Fig. 6).
An exploration of the reasons for heterogeneity rather than the computation of a single summary measure is an important goal of meta-analysis [45]. We found significant heterogeneity with regard to sensitivity, specificity, PLR, NLR, and DOR among the studies analyzed. In our meta-analysis, groups 2, and 3, and infection of the urinary tract substantially affected the sensitivity of the diagnosis of bacterial infection in the meta-regression analysis; Emergency Department (ED) and ICU affected the specificity of diagnosis of bacterial infection in the meta-regression analysis. In this meta-analysis, the studies were grouped according to Sackett and Haynes’ classification. In groups 1 and 3, the index test was developed in an ideal situation against a validation set (group 2), in which its performance was tested in a more realistic clinical context. Group 2 studies were the most informative for clinical practice as they were designed to resemble real-life situations by being restricted to patients who were most likely to be encountered by clinicians. And the test results by study design method of PR were interpreted more realistic than CR. The results of multiple meta-regression analyses showed that the sTREM-1 test had the highest accuracy for the diagnosis of bacterial infection in group 2 (sensitivity: 0.92, 95% CI 0.82, 0.97; specificity: 0.91, 95% CI 0.81, 0.96). The sensitivity of sTREM-1 for the diagnosis of bacterial infection was lowest in group 3. In addition, the study design method of PR + CR and PR showed better test performances than CR, especially PR + CR studies (sensitivity: 0.90, 95% CI 0.72, 0.79; specificity: 0.90, 95% CI 0.78, 0.96), potentially indicating the usefulness of measuring sTREM-1 in clinical practice. For the type of infection, the sTREM-1 test for diagnosis of urinary tract infection had low sensitivity (19%) and relatively high specificity (95%), possibly because inflammatory mediators such as sTREM-1 were diluted during urine production and removed from the bladder by diuresis, resulting in low sensitivity and a negative predictive value. For these reasons, the additive diagnostic value of sTREM-1 measurement in the urine of patients with a suspected UTI is limited [36]. This condition may also exist when testing sTREM-1 in other fluid-containing organs/tissues. For example, cerebrospinal fluid relatively high production may decrease sensitivity of the sTREM-1 test for diagnosis bacterial meningitis. And the sTREM-1 test for diagnosis bacterial infection in pleural cavity and blood had relatively high sensitivity. As a consequence, the threshold levels of sTREM-1 for the diagnosis of bacterial infection may vary between different organs/tissues.
Publication bias is common in diagnostic studies, possibly more so than in studies of randomized controlled trials [46]. We detected publication bias in our review. As expected, there was no significant publication bias in our review. However, the statistical methods used to assess publication bias have limitations [47–49]. Therefore, the above findings must be interpreted in this context.
Our meta-analysis had several limitations. First, the exclusion of conference abstracts and letters to the editors may have led to the publication bias that was observed in the present meta-analysis. Second, the number of eligible studies was small, and these studies dealt with very different things (such as study setting or assay method), potentially not reflecting the broader experience. An inflation of accuracy estimates could occur due to preferential acceptance of papers reporting favorable results. Third, some of the studies were small in number. Studies based on small samples sizes may have allowed for a Type II error. Because of the lack of required data reported in the original publications and the small number of eligible studies, we could not analyze the cutoff of the corresponding type of bacterial infection, and we also could not analyze the effects of factors such as laboratory infrastructure and patient spectrum (e.g., the severity of inflammation caused by infection) on the accuracy of sTREM-1 measurements.
Conclusions
The sTREM-1 assay appears to be a reliable marker of bacterial infection. Evaluations of sTREM-1 assays seem primarily to have been conducted with sample material from suspected sites of infection, but sTREM-1 may not be a sufficient biological marker for infection of the urinary tract due to its low sensitivity. The evidence presented in this review supports the usefulness of measuring sTREM-1 in clinical practice, except for the diagnosis of UTI. The real clinical value of the sTREM-1 assay is evident when sTREM-1 levels are low, allowing the clinician to withhold empiric antibiotics until culture results are available, thus eliminating unnecessary exposure of the patient to antibiotics. Whether sTREM-1 guidance can reduce antibiotic use will require additional studies, and additional prospective studies will be necessary to assess the usefulness of measuring sTREM-1 in different types of infection.
References
Hotchkiss RS, Karl IE (2003) The pathophysiology and treatment of sepsis. N Engl J Med 348:138–150
Davis BH (2005) Improved diagnostic approaches to infection/sepsis detection. Expert Rev Mol Diagn 5:193–207
Brun-Buisson C (2000) The epidemiology of the systemic inflammatory response. Intensive Care Med 26(Suppl 1):S64–S74
American College of Chest Physicians, Society of Critical Care Medicine Consensus Conference (1992) Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Crit Care Med 20:864–874
Kollef MH, Sherman G, Ward S, Fraser VJ (1999) Inadequate antimicrobial treatment of infections: a risk factor for hospital mortality among critically ill patients. Chest 115:462–474
Reinhart K, Meisner M, Brunkhorst FM (2006) Markers for sepsis diagnosis: what is useful? Crit Care Clin 22:503–519, ix–x
Ramirez P, Menendez R, Torres A (2007) New biological markers of ventilator-associated pneumonia. Clin Pulm Med 14:93–98
Gibot S, Kolopp-Sarda MN, Bene MC, Cravoisy A, Levy B, Faure GC, Bollaert PE (2004) Summaries for patients. A possible new test for diagnosing sepsis. Ann Intern Med 141:146
Jones AE, Fiechtl JF, Brown MD, Ballew JJ, Kline JA (2007) Procalcitonin test in the diagnosis of bacteremia: a meta-analysis. Ann Emerg Med 50:34–41
Tang BM, Eslick GD, Craig JC, McLean AS (2007) Accuracy of procalcitonin for sepsis diagnosis in critically ill patients: systematic review and meta-analysis. Lancet Infect Dis 7:210–217
Simon L, Gauvin F, Amre DK, Saint-Louis P, Lacroix J (2004) Serum procalcitonin and C-reactive protein levels as markers of bacterial infection: a systematic review and meta-analysis. Clin Infect Dis 39:206–217
Bouchon A, Dietrich J, Colonna M (2000) Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J Immunol 164:4991–4995
Tessarz AS, Cerwenka A (2008) The TREM-1/DAP12 pathway. Immunol Lett 116:111–116
Bleharski JR, Kiessler V, Buonsanti C, Sieling PA, Stenger S, Colonna M, Modlin RL (2003) A role for triggering receptor expressed on myeloid cells-1 in host defense during the early-induced and adaptive phases of the immune response. J Immunol 170:3812–3818
Bouchon A, Facchetti F, Weigand MA, Colonna M (2001) TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature 410:1103–1107
Deville WL, Buntinx F, Bouter LM, Montori VM, de Vet HC, van der Windt DA, Bezemer PD (2002) Conducting systematic reviews of diagnostic studies: didactic guidelines. BMC Med Res Methodol 2:9
Whiting P, Rutjes AW, Reitsma JB, Bossuyt PM, Kleijnen J (2003) The development of QUADAS: a tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med Res Methodol 3:25
Sackett D, Haynes R (2002) The architecture of diagnostic research. Br Med J 324(7336):539–541
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7:177–188
Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. Br Med J 315:629–634
Duval S, Tweedie R (2000) Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics 56:455–463
Tejera A, Santolaria F, Diez ML, Aleman-Valls MR, Gonzalez-Reimers E, Martinez-Riera A, Milena-Abril A (2007) Prognosis of community acquired pneumonia (CAP): value of triggering receptor expressed on myeloid cells-1 (TREM-1) and other mediators of the inflammatory response. Cytokine 38:117–123
Horonenko G, Hoyt JC, Robbins RA, Singarajah CU, Umar A, Pattengill J, Hayden JM (2007) Soluble triggering receptor expressed on myeloid cell-1 is increased in patients with ventilator-associated pneumonia: a preliminary report. Chest 132:58–63
Gibot S, Cravoisy A, Dupays R, Barraud D, Nace L, Levy B, Bollaert PE (2007) Combined measurement of procalcitonin and soluble TREM-1 in the diagnosis of nosocomial sepsis. Scand J Infect Dis 39:604–608
Liu CL, Hsieh WY, Wu CL, Kuo HT, Lu YT (2007) Triggering receptor expressed on myeloid cells-1 in pleural effusions: a marker of inflammatory disease. Respir Med 101:903–909
Phua J, Koay ESC, Zhang D, Tal LK, Boo XL, Lim KC, Lim TK (2006) Soluble triggering receptor expressed on myeloid cells-1 in acute respiratory infections. Eur Respir J 28:695–702
Richeldi L, Mariani M, Losi M, Maselli F, Corbetta L, Buonsanti C, Colonna M, Sinigaglia F, Panina-Bordignon P, Fabbri LM (2004) Triggering receptor expressed on myeloid cells: role in the diagnosis of lung infections. Eur Respir J 24:247–250
How CK, Chern CH, Wu MF, Wang LM, Huang CI, Lee CH, Hsieh SL (2007) Expression of the triggering receptor expressed on myeloid cells-1 mRNA in a heterogeneous infected population. Int J Clin Pract. doi:10.1111/j.1742-1241.2006.01193.x
El Solh AA, Akinnusi ME, Peter M, Berim I, Schultz MJ, Pineda L (2008) Triggering receptors expressed on myeloid cells in pulmonary aspiration syndromes. Intensive Care Med 34:1012–1019
Gibot S, Cravoisy A, Levy B, Bene MC, Faure G, Bollaert PE (2004) Soluble triggering receptor expressed on myeloid cells and the diagnosis of pneumonia. N Engl J Med 350:451–458
Gibot S, Kolopp-Sarda MN, Bene MC, Cravoisy A, Levy B, Faure GC, Bollaert PE (2004) Plasma level of a triggering receptor expressed on myeloid cells-1: its diagnostic accuracy in patients with suspected sepsis. Ann Intern Med 141:9–15
Determann RM, Millo JL, Gibot S, Korevaar JC, Vroom MB, van der Poll T, Garrard CS, Schultz MJ (2005) Serial changes in soluble triggering receptor expressed on myeloid cells in the lung during development of ventilator-associated pneumonia. Intensive Care Med 31:1495–1500
Determann RM, Weisfelt M, de Gans J, van der Ende A, Schultz MJ, van de Beek D (2006) Soluble triggering receptor expressed on myeloid cells 1: a biomarker for bacterial meningitis. Intensive Care Med 32:1243–1247
Bishara J, Hadari N, Shalita-Chesner M, Samra Z, Ofir O, Paul M, Peled N, Pitlik S, Molad Y (2007) Soluble triggering receptor expressed on myeloid cells-1 for distinguishing bacterial from aseptic meningitis in adults. Eur J Clin Microbiol Infect Dis 26:647–650
Chan MC, Chang KM, Chao WC, Lin LY, Kuo BI, Hsu JY, Wu CL (2007) Evaluation of a new inflammatory molecule (triggering receptor expressed on myeloid cells-1) in the diagnosis of pleural effusion. Respirology 12:333–338
Determann RM, Schultz MJ, Geerlings SE (2007) Soluble triggering receptor expressed on myeloid cells-1 is not a sufficient biological marker for infection of the urinary tract. J Infect 54:e249–e250
Kim JH, Park EY, Kim WH, Park W, Jeong HC, Lee JH, Kim EK (2007) Soluble triggering receptor expressed on myeloid cells-1: role in the diagnosis of pleural effusions. Tuberc Respir Dis 62:290–298
Kofoed K, Andersen O, Kronborg G, Tvede M, Petersen J, Eugen-Olsen J, Larsen K (2007) Use of plasma C-reactive protein, procalcitonin, neutrophils, macrophage migration inhibitory factor, soluble urokinase-type plasminogen activator receptor, and soluble triggering receptor expressed on myeloid cells-1 in combination to diagnose infections: a prospective study. Crit Care 11:R38
Yun SS, Jin HL, Eun MC, Jung HC (2007) Diagnostic utility of pleural fluid soluble triggering receptor expressed on myeloid cells 1 protein in patients with exudative pleural effusion. Tuberc Respir Dis 62:499–505
Chen HL, Hung CH, Tseng HI, Yang RC (2008) Soluble form of triggering receptor expressed on myeloid cells-1 (sTREM-1) as a diagnostic marker of serious bacterial infection in febrile infants less than three months of age. Jpn J Infect Dis 61:31–35
Huh JW, Lim CM, Koh Y, Oh YM, Shim TS, Lee SD, Kim WS, Kim DS, Kim WD, Hong SB (2008) Diagnostic utility of the soluble triggering receptor expressed on myeloid cells-1 in BAL fluid from patients with bilateral lung infiltrates. Crit Care 12:R6
Irwig L, Tosteson AN, Gatsonis C, Lau J, Colditz G, Chalmers TC, Mosteller F (1994) Guidelines for meta-analyses evaluating diagnostic tests. Ann Intern Med 120:667–676
Deeks JJ (2001) Systematic reviews in health care: systematic reviews of evaluations of diagnostic and screening tests. Br Med J 323:157–162
Wong-Baeza I, Gonzalez-Roldan N, Ferat-Osorio E, Esquivel-Callejas N, Aduna-Vicente R, Arriaga-Pizano L, Astudillo-de la Vega H, Villasis-Keever MA, Torres-Gonzalez R, Estrada-Garcia I, Lopez-Macias C, Isibasi A (2006) Triggering receptor expressed on myeloid cells (TREM-1) is regulated post-transcriptionally and its ligand is present in the sera of some septic patients. Clin Exp Immunol 145:448–455
Petitti DB (2001) Approaches to heterogeneity in meta-analysis. Stat Med 20:3625–3633
Irwig L, Macaskill P, Glasziou P, Fahey M (1995) Meta-analytic methods for diagnostic test accuracy. J Clin Epidemiol 48:119–130 Discussion 131–132
Stuck AE, Rubenstein LZ, Wieland D (1998) Bias in meta-analysis detected by a simple, graphical test. Asymmetry detected in funnel plot was probably due to true heterogeneity. Br Med J 316:469 Author reply 470–461
Vandenbroucke JP (1998) Bias in meta-analysis detected by a simple, graphical test. Experts’ views are still needed. Br Med J 316:469–470 Author reply 470–471
Song F, Eastwood AJ, Gilbody S, Duley L, Sutton AJ (2000) Publication and related biases. Health Technol Assess 4:1–115
Conflict of interest statement
The authors have no conflicts of interest or financial disclosures to declare.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Jiyong, J., Tiancha, H., Wei, C. et al. Diagnostic value of the soluble triggering receptor expressed on myeloid cells-1 in bacterial infection: a meta-analysis. Intensive Care Med 35, 587–595 (2009). https://doi.org/10.1007/s00134-008-1333-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00134-008-1333-z