Abstract
Objectives
We aimed to characterise and quantify the incidence of common infectious agents in acute exacerbations of chronic obstructive pulmonary disease (COPD) requiring ventilation, with a focus on respiratory viruses.
Design
An epidemiological study conducted over 3 years.
Setting
A 12-bed intensive care unit (ICU).
Participants
ICU patients over 45 years of age with a primary diagnosis of COPD exacerbation requiring non-invasive ventilation (NIV) or ventilation via endotracheal tube (ETT).
Materials and methods
Nasopharyngeal aspirates (NPA) and posterior pharyngeal swabs (PS) were tested for viruses with immunofluorescence assay (IFA), virus culture (VC) and polymerase chain reaction (PCR). Paired virus and atypical pneumonia serology assays were taken. Blood, sputum and endotracheal aspirates were cultured for bacteria.
Results
107 episodes in 105 patients were recorded. Twenty-three (21%) died within 28 days. A probable infectious aetiology was found in 69 patient episodes (64%). A virus was identified in 46 cases (43%), being the sole organism in 35 cases (33%) and part of a mixed infection in 11 cases (10%). A probable bacterial aetiology was found in 25 cases (23%). There was no statistically significant difference in clinical characteristics or outcomes between the group with virus infections and that without.
Conclusion
Forty-six (43%) of the patients with COPD exacerbation requiring mechanical ventilation had a probable viral pathogen. Prodromal, clinical and outcome parameters did not distinguish virus from non-virus illness. PCR was the most sensitive whilst virus culture was the least of virus assays.
Similar content being viewed by others
Introduction
Chronic obstructive pulmonary disease (COPD) is defined as the presence of progressive, incompletely reversible obstructive lung disease from diffuse causes, including chronic bronchitis, emphysema and small airways disease. Some 3–5% of approximately 1000 patients presenting annually with exacerbations of COPD are admitted to our intensive care unit (ICU) for ventilatory assistance with either bilevel non-invasive positive pressure ventilation (NIV) or ventilation via an endotracheal tube (ETT). Despite several studies in the unventilated population [1, 2, 3, 4, 5, 6], and two studies in ventilated COPD patients [7, 8], the viral aetiology of many COPD exacerbations remains unclear. Over 200 respiratory viruses [9] have been shown to cause upper and lower respiratory tract infections via inflammatory and neural mediators [10]. Of these, influenza types A and B (Inf A, B), parainfluenza types 1, 2 and 3 (Para 1, 2, 3), rhinovirus (RV), adenovirus (AV), respiratory syncytial virus (RSV), coronavirus (CoV) [11, 12] and, less commonly, human metapneumovirus (hMPV) [13], and enterovirus (EV) [14, 15] have been shown to play significant roles in airway infections.
We hypothesised that viral infections may be associated with a significant disease burden in patients with exacerbations of COPD admitted to ICU for ventilatory support. We aimed to investigate the incidence of these viruses. We also sought other known biological and atypical pathogens responsible for COPD exacerbations to build up as complete a microbiological profile as possible.
Materials and methods
Patient population
Any patient over the age of 45 years requiring mechanical ventilation for an exacerbation of COPD within 48 h of admission to hospital was considered for the study. These patients were admitted to the ICU for ventilatory support. We defined COPD as baseline FEV1 < 70% predicted with incompletely reversible obstructive spirometry as per American Thoracic Society/ European Respiratory (ATS/ERS) COPD consensus guidelines [16]. In the absence of prior spirometry, we assessed a smoking history of at least 20 pack years, physical examination, review of previous hospital records, chest radiograph changes suggestive of COPD, exclusion of other major co-morbidities as a primary cause of chronic breathlessness and chronic reduction in exercise tolerance to make the diagnosis of COPD. ICU admission for ventilatory support was considered when, despite optimal medical therapy and oxygen administration, there was acidosis (pH < 7.35), hypercapnia (PaCO2 > 45 mmHg), increasing respiratory distress and/or worsening fatigue or reduced consciousness [16].
The decision to mechanically ventilate was made by the attending ICU specialist based upon past history and current clinical and radiological information. Each patient was assessed for either NIV or endotracheal intubation using the guidelines of the British Thoracic Society [17]. Patients were excluded from the study who themselves elected not to have mechanical ventilation, or who were not mechanically ventilated because they were not expected to survive due to the severity of their underlying COPD or concurrent illnesses, after consultation with either themselves, if capable and conscious, or with their relatives/care givers. Patients with asthma completely reversible with bronchodilators, those less than 45 years old, those with restrictive lung disease or surgery or trauma within the previous 4 weeks and those admitted to general wards before ICU admission for more than 48 h (and therefore at risk of a hospital-acquired infection) were excluded. The presence of clinical pneumonia or the finding of chest radiograph changes did not exclude patients, as virus infections are well recognised to cause pneumonic changes [17].
Methods
The study was conducted between July 2000 and November 2003. Approval for the study was obtained from our hospital ethics committee and consent obtained from patients or their next-of-kin. The duration of prodromal symptoms of increased cough, worsening dyspnoea, increased quantity or discolouration of sputum or increased wheeze was estimated from the start of their symptomatic deterioration. Prior smoking history, exercise tolerance, baseline functional capability and the presence of other co-morbidities were recorded. Treatment prior to hospital admission was listed, including home oxygen, corticosteroid therapy and antibiotics in the previous 3 days. Baseline clinical parameters, blood results, APACHE II score and arterial blood gas prior to commencement of mechanical ventilation were noted.
Every effort was made to exclude other causes of respiratory failure, such as respiratory depressants, uncontrolled oxygen therapy, pulmonary embolism, increased atmospheric pollution, acute left ventricular failure and pneumothorax by using careful history taking, clinical acumen, radiology, angiography and echocardiography where appropriate.
Full blood count, chemistry, blood cultures and chest radiography were performed on admission to hospital. Sputum specimens were collected on admission, if possible, or within 48 h of hospital admission, and viral nasal and pharyngeal specimens were taken after patient stabilisation and enrolment in the study. Sputum for bacterial analysis was collected via endotracheal tube suction in intubated patients and by spontaneous or voluntary cough from patients who received NIV. All other specimen collection was identical.
Nasopharyngeal aspirate (NPA) and posterior pharyngeal swab (PS) (Copan Venturi Transystem, Italy) were collected from all patients. Immunofluorescence assay (IFA), either direct (Merifluor Bioscience, Cincinnati, Ohio) or indirect (Chemicon International, Temecula, CA), and virus culture (VC) were performed on the NPA. Virus culture only was performed on the PS and polymerase chain reaction (PCR) on both specimens. All patients were routinely treated with bronchodilators and empirical antibiotics for at least 5 days, changed or ceased according to the subsequent microbiological profile. All patients received physiotherapy when clinically stable.
New chest radiograph changes on admission implied pneumonic change or collapse which was not present on previous chest films (if available) or which was clearly abnormal and had the hallmarks of a primary pulmonary pathological process. Whether changes were new or chronic was agreed by consensus between a thoracic physician and an intensivist.
Microbiological interpretation
We have defined a probable pathogen as “the most likely biological agent(s) to have caused the exacerbation of COPD”. Acute serological titres for Inf A and B, Para 1, 2 and 3, RSV, AV, Chlamydia, Legionella and Mycoplasma antibodies were followed 3 weeks later with a paired convalescent titre. A fourfold rise in titre signified a probable causative agent. Virus nucleic acid extraction with or without reverse transcriptase and amplification was performed on NPA and PS samples. PCR detection was performed for Inf A and B, Para 1, 2 and 3 and RSV types A and B (Hexaplex assay) and human EV, hMPV, CoV, RV and AV (real-time PCR). Appendix 1 in the Electronic Supplementary Material (ESM) contains further details. A positive PCR viral assay was considered evidence for a probable virus pathogen.
A screening genus-specific Chlamydia assay (both IgG and IgA) was initially performed. For the purpose of this study, seroconversion of either or both IgG and IgA between acute and convalescent sera was considered evidence of acute Chlamydia genus infection. A single acute or convalescent specimen with either IgA, or IgA and IgG present was indicative of probable recent infection with Chlamydia. A positive IgG without IgA was not considered evidence of recent Chlamydia infection.
Sputum processing was performed using standard laboratory media. Microscopy of Gram stain smears as well as identification of all significant isolates was reported. Sputum samples were considered non-representative of a lower respiratory tract specimen if there were fewer than 20 neutrophils per high-power field or more than 25 squamous epithelial cells per low-power field. The significance of a culture result was based on the quality of the sputum and whether the culture matched the smear result [18]. Identified bacteria were defined as probable pathogens if the predominant growth of a potential respiratory pathogen was obtained in the presence of Gram stain microscopy showing profuse organisms with a consistent morphotype. Other bacterial isolates were considered as commensals or improbable pathogens.
Statistical analysis
Statistical analysis was performed using R (version 2.0.1) (www.r-project.org). Categorical variables were analysed using the chi-squared test or Fisher's exact test as appropriate, depending on whether numbers in the contingency tables satisfied the chi-squared test assumptions. Continuous variables were analysed using the Mann–Whitney U-test for non-normally distributed variables or the unpaired t-test for normally distributed variables. A p value of < 0.05 was considered significant.
Results
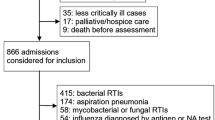
Of 118 potential participants (120 potential episodes), a total of 13 were not entered in the study for the following reasons: participation was declined (4); death before inclusion (6); enrolment more than 48 h after hospital admission (2); or failure to meet the criteria for the diagnosis of COPD (1). Two patients were admitted twice with separate episodes. Three patients were included in this study without hypercarbia > 45 mmHg. The indications for NIV in these cases were a combination of hypoxia (PO2 < 55) in spite of adequate oxygen therapy, pH < 7.35 and symptomatic relief. Of the 105 patients (107 episodes), 23 (21%) died within 28 days of admission. The mean APACHE II score was 20 (range 9–40) (Table 1). Less than 40% of the patients had recent or any pre-morbid spirometry values, and these data were therefore omitted in reporting. Convalescent serological follow-up of initial viral and atypical titres was 52%. We were unable to obtain a sputum specimen suitable for bacterial analysis in 27/107 patient episodes (25%). All these patients received NIV.
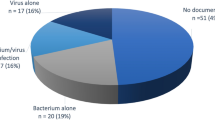
A probable infectious aetiology was found in 69 episodes (64%). A virus was the probable pathogen in 46 cases (43%), being the sole organism in 35 cases (33%) (Table 2) and part of a mixed infection in 11 cases (10%) (Table 3). Virus cultures for herpes simplex virus type 1 (HSV1) were positive in 8 cases (7%). Six HSV1 cases also had another probable pathogen identified and the remaining two had no distinguishable clinical or radiological features that identified HSV as a probable pathogen. These eight HSV1 results were therefore not included as probable pathogens.
Only two other viral cultures were positive, one for Inf A and one for RV, both of which correlated with a positive PCR. Inf A was the most common organism in 14 cases (13%) with 7 (8%) RV, 3 (3%) CoV, 3 (3%) hMPV and 2 (2%) EV. Twenty-four (22%) of 107 episodes occurred in the Australian summer and autumn months from December to May, whilst the bulk of exacerbations, 83/107 (78%), occurred in the winter and spring months between June and November (p = 0.0000007). Figure 1 shows the seasonal breakdown.
There were two discrete clusters of influenza A that occurred each within a 3-week period, both in early spring and separated by 3 years, and comprising 9/14 (65%) of the influenza A-positive cases. There was no pattern of clustering in any of the other infection types.
A probable bacterial aetiology was found in 25 cases (23%) (Table 2), being a sole agent in 16 cases (15%) and in a mixed infection in a further 9 (8%) (Table 3). Haemophilus influenzae was the most common bacterium in10/107 cases (11%). Chlamydia species were the only atypical organisms found, being the sole aetiology in 3 cases (3%) and in 3 mixed infections (3%). Mixed infections constituted 13 (12%) of all cases (Table 3).
There was no statistically significant difference between the group with probable viral aetiology and those without a viral aetiology in length of prodrome, initial clinical parameters, APACHE II score, changes on chest radiograph, length of stay or mortality (Table 4).
Discussion
There are only two other published series of infective agents in ventilated COPD patients [7, 8]. A subsequent study also examined the outcomes of treatment in one of these initial series [19]. All 50 patients in the Soler study [7] and all 54 in the Fagon study [8] were intubated, facilitating protected brush and bronchoalveolar lavage specimens. No bronchoscopic specimen collection was performed on the smaller subset of intubated patients in our study, where 61 patients (57%) received NIV alone, with 17 (16%) receiving both NIV and ETT and only 29 (27%) being intubated without NIV. The collection of appropriate respiratory samples has been complicated by the introduction of NIV as an accepted modality for the treatment of severe COPD exacerbations, as NIV patients may not produce suitable sputum. Fibre-optic bronchoscopy may not be easily tolerated by these patients, in spite of the use of modified face masks [20]. Inducement of sputum specimens with hypertonic saline, although an effective method of sample collection in unintubated patients [21, 22], may induce or worsen bronchospasm and was not considered a practicable option in our cohort with initial respiratory instability.
Bacteria grown from sputum or endotracheal aspirate may not be representative of the pathogenic organism that is causing the exacerbation [23, 24, 25], with up to half of stable COPD patients colonised by a potential bacterial pathogen [26, 27]. To reduce this possibility, we examined our sputum specimens for the presence of neutrophils and correlated the Gram stain with culture in an effort to determine whether the organism was a commensal or a probable pathogen [18]. Twenty-five (24%) of the 105 patients received antibiotics within the 3 days prior to ICU admission. Of these 25, 3 (12%) had a positive bacterial culture (all H. influenzae). The possibility that recent antibiotic therapy may have reduced the bacterial yield in the other 22 episodes must be considered.
Both Soler [7] and Fagon [8] focused primarily on the bacterial yield. Fagon did not investigate for viruses. Soler obtained serology for Chlamydia, Mycoplasma, Legionella, Inf, Para, RSV and HSV. AV, hMPV, CoV, EV and RV were not sought. Soler found potential pathogens in 72% of cases, compared with 64% in our study, and community-acquired bacteria in 56% of cases in his cohort, against 23% in our study. Fagon found bacteria in 27/54 (50%) of protected brush specimens, predominantly Haemophilus and Streptococcus species. Soler did not find significant clinical or outcome differences between the bacteria-positive and -negative groups, whilst Fagon noted significantly greater pyrexia on presentation in the bacteria-positive group, but in no other parameters.
Protected brush specimens may well increase the overall yield of bacteria, whilst ensuring the false-positive rate is minimised.
The limited range of viruses sought and the lack of PCR assay may well have underrepresented virus incidence (16%) in Soler's study, compared with 43% in ours. He found atypical organisms in 18% of cases, compared with our 5%. With adequate follow-up serology in only 67% of the survivors in our study, this may account for the paucity of proven atypical organisms. Some 33% of cases were polymicrobial in Soler's study, compared with 13% in ours.
The mechanisms for viral upper respiratory tract infections (URTIs) causing lower respiratory tract infection (LRTI) symptoms include cytokine and paracrine release, altered cell-mediated immunity and lymphocyte recruitment [28], neurohormonal reflexes and direct epithelial damage of the lower respiratory tract [10, 29, 30, 31]. We may therefore assume that the recovery of common viruses from the upper respiratory tract samples (NPA and PS) reflects a probable viral aetiology for lower respiratory tract symptoms. A lower sensitivity for viral diagnosis than for direct bronchoscopic sampling of lower respiratory tract specimens is possible [22, 32, 33].
A virus aetiology has been implicated in non-ventilated COPD exacerbations in between 29 and 64% of cases [6, 32, 34, 35], with the most common virus detected in each series varying due to seasonal variation, method of virus detection and influenza vaccination status. InfA [6, 36], RSV [32, 33, 36], CoV [37] and RV [5, 22, 32, 33] were the most frequently detected viral pathogens in more recent studies of COPD exacerbations. In our study, Inf A, Para 3, RSV and RV were most often detected (Table 2). One study suggests that patients with evidence of virus infection during COPD exacerbations have greater symptom severity at onset than those with non-virus exacerbations, and a significantly longer median time to symptom recovery [32]. We did not find this in our study, with no significant differences in prodromal symptoms, length of ICU and hospital stay or mortality observed between virus and non-virus exacerbations.
Our study is the first to use PCR to detect viruses in ventilated COPD patients. PCR was the most sensitive method for virus detection, with virus culture the least sensitive. Garbino reports a 10% virus culture positivity from bronchoalveolar lavage (BAL) specimens in a group of hospitalised patients with LRTIs, whilst reverse transcriptase PCR (RT-PCR) identified a virus in 29% of the same specimens [33]. This may be because the presence of only small quantities of virus is not detectable by culture. Most viral pathogens are more difficult to culture after more than 72 h of illness and PCR is able to detect infection for a longer period due to its high sensitivity. PCR is consistently associated with high detection rates in other studies of the COPD exacerbation population, with a 40--64% virus yield [5, 6, 32]. A rising antibody titre over 3 weeks is considered strong evidence for a probable virus or atypical pathogen, but is obviously not possible in deceased patients or those lost to follow-up. Whilst a useful epidemiological tool, it provides a retrospective diagnosis and therefore cannot affect clinical decisions in the critically unwell.
We used new genetic techniques to enhance the diagnostic yield and provide a more accurate picture of the role of viral infection (see Appendix 1, ESM). However, most cases were diagnosed by RT-PCR alone. This heavy reliance on RT-PCR to establish a diagnosis of viral infection may be too sensitive, detecting small amounts of residual viral nucleic acid without other clinical evidence of viral infection. Extensive precautions were taken against contamination in our study, and negative PCR controls showed no evidence of false positivity.
Respiratory viruses may be commensal in stable COPD patients. Seemungal found respiratory viruses more often in the sputum and nasal lavage of patients with exacerbations of COPD (56%) than of patients with stable COPD (19%) [6, 32]. Latent sequences of common respiratory viruses such as adenovirus [5, 37] have been detected by PCR. It has been well demonstrated that COPD patients may excrete RSV for extended periods of time [19], so one cannot be absolutely certain that the RSV demonstrated in our study was necessarily pathogenic. For other viruses, persistent colonisation is not well described and it is reasonable to assume that they played a pathogenic role.
Quantitative real-time PCR may allow differentiation between latent or former infection and current infection. Advances in the availability and rapidity of molecular sequencing may also allow changes in infectious agent serotype or antibiotic resistance to be discerned [33, 38].
There is no evidence to date to suggest that treatment with antiviral agents in this subset of COPD patients with virus exacerbations requiring ventilatory support will significantly alter the course of their acute illness. It is nevertheless important to have a better understanding of the pattern of disease in order to target more efficiently any potential treatments. The early detection of virus infection by PCR is in its infancy, but the development of more sophisticated anti-viral agents may allow early targeting of virus pathogens.
Conclusions
A probable virus pathogen was found in 46 cases (43%) and a probable bacterial aetiology was found in 25 cases (23%) in this study of ventilated COPD exacerbation patients. No prodromal or initial clinical parameters distinguished viral from non-viral illness in our study. There were no significant differences between virus and non-virus exacerbations in ventilation time/length of stay or 28-day mortality. NIV made protected specimen collection more difficult than in intubated patients, which may have decreased the bacterial and viral yield. PCR offered a more detailed view of the spectrum of virus illness in this group of patients, including previously unsought viruses such as EV and hMPV.
References
Walsh E, Falsey A, Hennessey P (1999) Respiratory syncytial and other virus infections in persons with chronic cardiopulmonary disease. Am J Resp Crit Care Med 160:791–795
Upshur R, Knight K, Goel V (1999) Time series analysis of the relation between influenza virus and hospital admissions of the elderly in Ontario, Canada, for pneumonia, chronic lung disease, and congestive heart failure. Am J Epidem 149:85–92
Smith C, Kanner R, Golden C, Klauber M, Renzetti A Jr (1980) Effect of viral infections in patients with chronic obstructive pulmonary diseases. J Infect Dis 141:271–280
Lowenburg A, Orie N, Sluiter H, de Vries K (1986) Bronchial hyperreactivity and bronchial obstruction in respiratory viral infection. An attempt to evaluate the relationship. Respiration 49:1–9
Bandi V, Jakubowycz M, Kinyon C, Mason EO, Atmar RL, Greenberg SB, Murphy TF (2003) Infectious exacerbations of chronic obstructive pulmonary disease associated with respiratory viruses and non-typeable Haemophilus influenzae. FEMS Immunol Med Microbiol 37:69–75
Rohde G, Wiethege A, Borg I, Kauth M, Bauer TT, Gillissen A, Bufe A, Schultze-Werninghaus G (2003) Respiratory viruses in exacerbations of chronic obstructive pulmonary disease requiring hospitalisation: a case-control study. Thorax 58:37–42
Soler N, Torres A, Ewig S, Gonzalez J, Celis R, El-Ebiary M, Hernandez C, Rodriguez-Roisin R (1998) Bronchial microbial patterns in severe exacerbations of chronic obstructive pulmonary disease (COPD) requiring mechanical ventilation. Am J Respir Crit Care Med 157:1498–1505
Fagon JY, Chastre J, Trouillet JL, Domart Y, Dombret MC, Bornet M, Gibert C (1990) Characterization of distal bronchial microflora during acute exacerbation of chronic bronchitis. Use of the protected specimen brush technique in 54 mechanically ventilated patients. Am Rev Respir Dis 142:1004–1008
Mackie PL (2003) The classification of viruses infecting the respiratory tract. Paediatric Respiratory Reviews 42:84–90
Folkerts G, Nijkamp FP (1995) Virus-induced airway hyperresponsiveness. Role of inflammatory cells and mediators. Am J Respir Crit Care Med 151:1666–1673
McKean MC, Leech M, Lambert PC, Hewitt C, Myint S, Silverman M (2001) A model of viral wheeze in nonasthmatic adults: symptoms and physiology. Eur Respir J 18:23–32
Jacoby DB (2002) Virus-induced asthma attacks. JAMA 287:755–761
Williams JV, Harris PA, Tollefson SJ, Halburnt-Rush LL, Pingsterhaus JM, Edwards KM, Wright PF, Crowe JE Jr (2004) Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N Engl J Med 350:443–450
Jartti T, Lehtinen P, Vuorinen T, Osterback R, van den Hoogen B, Osterhaus AD, Ruuskanen O (2004) Respiratory picornaviruses and respiratory syncytial virus as causative agents of acute expiratory wheezing in children. Emerg Infect Dis 10:1095–1099
Knipe D, Howley P (2001) Field's virology, 4th edn. Lippincott Williams & Wilkins, ch 24, p 755
Celli BR, MacNee W, Force AET (2004) Standards for the diagnosis and treatment of patients with COPD: a summary of the ATS/ERS position paper. Eur Respir J 23:932–946
Committee, BTSSoC (2002) Non-invasive ventilation in acute respiratory failure. Thorax 57:192–211
Eisenberg H (2004) Aerobic bacteriology – respiratory tract cultures. Clinical Microbiology Procedures Handbook, 2nd edn. ASM Press, Washington, DC
Ewig S, Soler N, Gonzalez J, Celis R, El-Ebiary M, Torres A (2000) Evaluation of antimicrobial treatment in mechanically ventilated patients with severe chronic obstructive pulmonary disease exacerbations. Crit Care Med 28:692–697
Antonelli M, Conti G, Riccioni L, Meduri G (1996) Noninvasive positive-pressure ventilation via face mask during bronchoscopy with BAL in high-risk hypoxemic patients. Chest 110:724–728
Borg I, Rohde G, Loseke S, Bittscheidt J, Schultze-Werninghaus G, Stephan V, Bufe A (2003) Evaluation of a quantitative real-time PCR for the detection of respiratory syncytial virus in pulmonary diseases. Eur Respir J 21:944–951
Seemungal TA, Harper-Owen R, Bhowmik A, Jeffries DJ, Wedzicha JA (2000) Detection of rhinovirus in induced sputum at exacerbation of chronic obstructive pulmonary disease. Eur Respir J 16:677–683
Chitkara RK, Sarinas PS (2002) Recent advances in diagnosis and management of chronic bronchitis and emphysema. Curr Opin Pulm Med 8:126–136
Soto FJ, Varkey B (2003) Evidence-based approach to acute exacerbations of COPD. Curr Opin Pulm Med 9:117–124
Miravitlles M (2002) Exacerbations of chronic obstructive pulmonary disease: when are bacteria important? Eur Respir J [Suppl] 36:9s–19s
Banerjee D, Khair OA, Honeybourne D (2004) Impact of sputum bacteria on airway inflammation and health status in clinical stable COPD. Eur Respir J 23:685–691
Patel IS, Seemungal TA, Wilks M, Lloyd-Owen SJ, Donaldson GC, Wedzicha JA (2002) Relationship between bacterial colonisation and the frequency, character, and severity of COPD exacerbations. Thorax 57:759–764
Johnston SL (1995) Natural and experimental rhinovirus infections of the lower respiratory tract. Am J Respir Crit Care Med 152:S46–S52
Sterk PJ (1993) Virus-induced airway hyperresponsiveness in man. Eur Respir J 6:894–902
Johnston SL, Papi A, Bates PJ, Mastronarde JG, Monick MM, Hunninghake GW (1998) Low grade rhinovirus infection induces a prolonged release of IL-8 in pulmonary epithelium. J Immunol 160:6172–6181
Gern JE, Galagan DM, Jarjour NN, Dick EC, Busse WW (1997) Detection of rhinovirus RNA in lower airway cells during experimentally induced infection. Am J Respir Crit Care Med 155:1159–1161
Seemungal T, Harper-Owen R, Bhowmik A, Moric I, Sanderson G, Message S, Maccallum P, Meade TW, Jeffries DJ, Johnston SL, Wedzicha JA (2001) Respiratory viruses, symptoms, and inflammatory markers in acute exacerbations and stable chronic obstructive pulmonary disease. Am J Respir Crit Care Med 164:1618–1623
Garbino J, Gerbase MW, Wunderli W, Deffernez C, Thomas Y, Rochat T, Ninet B, Schrenzel J, Yerly S, Perrin L, Soccal PM, Nicod L (2004) Lower respiratory viral illnesses: improved diagnosis by molecular methods and clinical impact. Am J Respir Crit Care Med 170:1197–1203
Glezen WP, Greenberg SB, Atmar RL, Piedra PA, Couch RB (2000) Impact of respiratory virus infections on persons with chronic underlying conditions. JAMA 283:499–505
Ball P (1995) Epidemiology and treatment of chronic bronchitis and its exacerbations. Chest 108:43S–52S
Greenberg SB (2002) Respiratory viral infections in adults. Curr Opin Pulm Med 8:201–208
Greenberg SB, Allen M, Wilson J, Atmar RL (2000) Respiratory viral infections in adults with and without chronic obstructive pulmonary disease. Am J Respir Crit Care Med 162:167–173
Nijhuis M, van Maarseveen N, Schuurman R, Verkuijlen S, de Vos M, Hendriksen K, van Loon AM (2002) Rapid and sensitive routine detection of all members of the genus enterovirus in different clinical specimens by real-time PCR. J Clin Microbiol40 10:3666–3670
Author information
Authors and Affiliations
Corresponding author
Additional information
The study was self-funded by internal grants from the Central Coast Health Services Research Fund and the Hunter Area Pathology Service. There was no funding from pharmaceutical or other commercial organisations or agencies.
There was no conflict of interest.
This article is discussed in the editorial available at: http://dx.doi.org/10.1007/s00134-006-203-9
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Cameron, R.J., de Wit, D., Welsh, T.N. et al. Virus infection in exacerbations of chronic obstructive pulmonary disease requiring ventilation. Intensive Care Med 32, 1022–1029 (2006). https://doi.org/10.1007/s00134-006-0202-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00134-006-0202-x