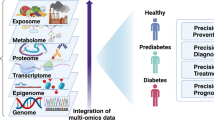
Abstract
Epidemiologists have for many decades reported on the patterns and distributions of diabetes within and between populations and have helped to elucidate the aetiology of the disease. This has helped raise awareness of the tremendous burden the disease places on individuals and societies; it has also identified key risk factors that have become the focus of diabetes prevention trials and helped shape public health recommendations. Recent developments in affordable high-throughput genetic and molecular phenotyping technologies have driven the emergence of a new type of epidemiology with a more mechanistic focus than ever before. Studies employing these technologies have identified gene variants or causal loci, and linked these to other omics data that help define the molecular processes mediating the effects of genetic variation in the expression of clinical phenotypes. The scale of these epidemiological studies is rapidly growing; a trend that is set to continue as the public and private sectors invest heavily in omics data generation. Many are banking on this massive volume of diverse molecular data for breakthroughs in drug discovery and predicting sensitivity to risk factors, response to therapies and susceptibility to diabetes complications, as well as the development of disease-monitoring tools and surrogate outcomes. To realise these possibilities, it is essential that omics technologies are applied to well-designed epidemiological studies and that the emerging data are carefully analysed and interpreted. One might view this as next-generation epidemiology, where complex high-dimensionality data analysis approaches will need to be blended with many of the core principles of epidemiological research. In this article, we review the literature on omics in diabetes epidemiology and discuss how this field is evolving.

Graphical abstract
Similar content being viewed by others
Avoid common mistakes on your manuscript.



Introduction
The aetiology and clinical presentation of diabetes often differ greatly from one patient to the next, as do patients’ responses to therapies and the rates at which they develop complications. Identifying biomarkers that aid the prediction and prevention of diabetes by helping stratify populations depending on (1) sensitivity to risk factors, (2) likely response to therapies, and (3) susceptibility to diabetes complications, as well as identifying biomarkers for disease monitoring and as surrogate outcomes, are major priorities in diabetes research.
Biomarkers are also used extensively in diabetes epidemiology as intermediate exposure or outcome variables when seeking to understand disease aetiology. For example, HbA1c and blood glucose concentrations are the principal biomarkers of diabetes, and measures of blood concentrations of insulin, proinsulin, lipids, inflammatory cytokines and adipokines are often used to study the determinants or consequences of diabetes.
The development of high-throughput molecular genotyping and phenotyping assays has led to a new field of omics research, which has seen the discovery of many types of biological variants influencing diabetes. This review explores diabetes epidemiology, with specific focus on omics research. How the next generation of epidemiological studies and methods are likely to evolve and contribute to understanding diabetes is also discussed.
What are biomarkers?
The term ‘biomarker’ was first used in the field of petroleum chemistry in the late 1960s [1], appearing a few years later in the biomedical literature to describe the role of serum RNase as an indicator of renal function [2]. The National Institutes of Health’s Biomarkers Definitions Working Group subsequently defined a biomarker as ‘A characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention’ [3]; while other definitions have followed, this early one remains widely used. Biomarkers have multiple uses in biomedicine, including in drug trials, though discussion of their use in such trials is outside the remit of this article. Nevertheless, the US Food and Drug Administration’s (FDA) ‘context of use’ framework for the use of biomarkers in drug trials provides a reasonable foundation upon which biomarkers in many areas of epidemiology research can be considered. Briefly, the FDA cites seven specific contexts within which biomarkers can be used in drug trials: (1) diagnosis (for patient selection), (2) monitoring (disease development, toxicity, exposure), (3) prediction (effects of interventions or exposures), (4) prognosis (patient stratification and/or enrichment), (5) pharmacodynamics/response (efficacy: surrogate endpoints and/or biological response to treatment), (6) safety, and (7) susceptibility/risk (potential to develop disease or exposure sensitivity) (see www.fda.gov/drugs/cder-biomarker-qualification-program/context-use) [4, 5]. An extended overview is provided in the Text box ‘Context of use of biomarkers’ [6].
The evolution of omics in epidemiology
Comprehensive molecular phenotyping in very large cohort collections has facilitated the discovery of many previously unknown biological pathways, providing substrates for drug development pipelines, the optimisation of non-pharmacological interventions, disease-monitoring technologies and disease-prediction algorithms. This has involved genome-wide association studies (GWAS) and their statistical aggregation through meta-analysis [7].
GWAS usually require large cohort collections to afford adequate power, mainly because many parallel hypotheses are tested (>1 million) and risk of type 1 error (false-positive discovery) is consequently high. A limitation of GWAS is the inability to detect certain types of variants, either because they were absent within the populations used to inform the content of GWAS arrays or imputation panels, or because the array is simply not designed to detect certain types of variant. This can prove problematic when studying rare variants [8], but also applies to non-SNP variants such as insertion–deletion polymorphisms (indels) [9, 10], although this may be less of a concern than initially thought [8]. Alternatively, whole-genome sequencing involves the interrogation of each accessible base pair in the nuclear genome in a manner that is largely agnostic to the identity of the specific variants. Thus, with sequencing, previously unknown variants (or at least those not included in genotyping arrays) can be discovered and related to phenotypic variation. As an example, homozygote carriers of the TBC1D4 nonsense p.Arg684ter allele, common in the Greenland Inuit population but rare elsewhere, have ~10-fold increased odds of type 2 diabetes [11]. This causal variant is tagged by genotypes captured in certain arrays, but the causal variant itself is not captured; thus, its detection required de novo exome sequencing of DNA from Inuit trios (mother, father and a child).
Many other types of omics data (e.g. transcriptomics, proteomics, microRNAs, epigenetics, peptidomics, metabolomics, lipidomics, metagenomics) can also be derived from stored biosamples using targeted assays, arrays or sequencing technologies, depending on storage procedures [12] (see Table 1, and Fig. 1 and Text box: Potential challenges during the retrieval of omics data).
Omics studies workflow. Initial stages of omic studies involve the ethical approval of the study protocol (research ethics) and written consent (participant recruitment) provided by the participants where biological samples are drawn for further analyses. Downstream stages include critical steps, i.e. sample storage and processing, data generation, and data analysis (integration, interpretation and dissemination). This figure is available as a downloadable slide
Epidemiology and its role in diabetes research
Epidemiology, the study of disease, its risk factors and its consequences within human populations, has been a cardinal feature of diabetes research for almost a century. In the Whitehall II Study, for example, 6538 British civil servants, initially free from diabetes, were studied repeatedly for about a decade [13]. An analysis of these data showed that over the decade preceding type 2 diabetes diagnosis, fasting and post-load blood glucose concentrations gradually increased, deteriorating sharply in the final 3 years. Compensatory changes in estimated insulin production and insulin sensitivity also occurred, whereby insulin sensitivity declined rapidly during the final 5 years before diagnosis and insulin production initially increased from years four to three pre-diagnosis, only to decline rapidly thereafter. In those who did not develop diabetes, fasting blood glucose concentrations and insulin production remained materially unchanged throughout follow-up, whereas post-load glucose rose gradually, and insulin sensitivity declined throughout follow-up at rates similar to those seen in participants who developed diabetes.
‘Correlation’ does not necessarily mean ‘causation’; some types of epidemiological analyses, such as those focused on prediction (for example, of risk of developing diabetes, of susceptibility to risk factors or of treatment success/failure) do not always require that the relationships between exposures and outcomes are causal for the results to be clinically useful. Similarly, descriptive epidemiology does not seek to establish cause and effect, instead focusing on detailing the patterns and distributions of disease. However, in aetiological epidemiology, where attention is often placed on understanding mechanisms of action, establishing causality is paramount, especially where focus is on discovery of novel drug targets that perturb pathways influencing diabetes or diabetes complications.
The major barriers to causal inference in epidemiology are chance, bias and confounding. These challenges can be addressed to some extent by applying certain data analysis conventions, such as the Bradford Hill criteria [14] (see Text box: Bradford Hill criteria in next-generation epidemiology). Of the many quantitative approaches for causal inference, Mendelian randomisation (MR), which often utilises genetic variants as the ‘causal instrument’, is popular. SNPs, unlike most other types of biomarker, remain constant throughout life. Thus, unlike most other biomarkers, there is no need to reassess genotypes once on file. This stability also means that cross-sectional associations between genotypes and traits can be considered unidirectional. MR exploits these strengths as well as the random assortment of genotypes to minimise the impact of confounding and reverse causality [15].
Several branched-chain amino acids (BCAAs) such as isoleucine, leucine and valine have been among the ~100 biomarkers reproducibly associated with type 2 diabetes incidence in large observational studies [16]. Of these, alanine aminotransferase, proinsulin and uric acid are also supported by causal evidence from MR studies [17]. Early studies exploring the causal link between vitamin D and diabetes, using a genetic instrument comprised of variants associated with circulating levels, showed conflicting evidence [18,19,20]. However, in later studies, when the sample size and the instrumental variables were expanded and the genetic instrument included variants regulating the synthesis, transport and catabolism of vitamin D, a causal relationship was evident [21].
Most MR studies have focused on prevalent diabetes, with relatively few (about ten) biomarkers being causally associated with incident type 2 diabetes [22]. A recent elegant analysis [23] of biomarkers in incident diabetes reported that 35 biomarkers have been studied in cohorts totalling at least 1000 individuals with type 2 diabetes, only one of which (ferritin) yielded strong observational and MR evidence to support a causal role in diabetes incidence. In general, the biomarkers examined did not enhance the accuracy of type 2 diabetes prediction models, and those that did were generally markers of glycaemic control.
Although MR is often viewed as a highly robust means of inferring causal relationships, the approach has noteworthy caveats [24]. For example, Haworth and colleagues [25] describe geographically aligned genetic structures associated with traits such educational attainment, BMI and number of siblings, using data from the UK Biobank, which raises concerns about the validity of some published MR analyses.
The value of biobanked samples and longitudinal cohorts
Biorepositories have existed for several decades, although the term ‘biobank’ was first used in the late 1990s [26]. The manner in which biobanks would be used today could not have been known when they were first initiated. Nevertheless, modern genotyping and phenotyping technologies have helped raise the value of many biobanks that were initiated long before these technologies were invented. In the UK, the Department of Health, the Medical Research Council, the Scottish Executive, and the Wellcome Trust invested UK£62m to establish UK Biobank, a prospective cohort study of 500,000 adults from the UK. Roughly 5% of the cohort has prevalent or incident diabetes [27], representing a large case group that is set to expand as the cohort ages. Established as a non-profit project in the early 2000s, UK Biobank has proved to be an outstanding resource for aetiological epidemiology owing to the extensive genotyping, relatively deep phenotyping and linkage with electronic health records. The thousand or so papers published in the past 7 years using UK Biobank data have spanned many health topics, with several dozen papers relating explicitly to diabetes. A common criticism of biobank research is that many are too small to stand alone and have thus formed parts of larger biobank networks, where data harmonisation has been challenging owing to the variety of methods deployed to assess the same underlying exposures and outcomes. Thus, as a single, large, standardised bioresource, UK Biobank has helped to address this criticism.
Next-generation epidemiology
The idea of genotyping and repeatedly phenotyping the same individual using multiple omics platforms was stimulated by a study in a single adult man who underwent deep omics profiling (genomics, transcriptomics, proteomics, metabolomics and autoantibodies) once daily for 14 months [28]. This analysis provided evidence that by integrating dense personal omics data, temporal patterns could be identified to predict subsequent shifts in health and disease markers. While the ‘n of 1’ nature of this study limits its generalisability, the technical approaches deployed inspired others to undertake epidemiological studies involving deep phenotyping of existing biosamples, as well as new studies where participants were repeatedly assessed using digital and serological assays to profile temporal patterns related to the development or progression of diabetes.
Applying modern molecular phenotyping technologies to samples stored from historical cohorts is highly pragmatic, particularly when the cohort has a long follow-up and clinical events have accrued. The European Prospective Investigation into Cancer and Nutrition (EPIC)-InterAct (n = 12,500 incident cases and n = 16,000 reference cohort) is one of the largest nested case–cohort studies of incident diabetes. The study comprised subgroups of participants identified from a larger European prospective cohort study (EPIC, N = 500,000). The aim of InterAct was to assess gene–lifestyle interactions, but it has subsequently been used to address many other questions, including those focused on the role of diet in diabetes. Among the biomarkers analysed were plasma phospholipid fatty acids by gas chromatography. Imamura et al [29] used these data to derive a dietary fatty acid score, which they found to be inversely related to incident diabetes. In post hoc analyses, the same score was inversely associated with higher levels of liver enzymes, inflammatory markers, fasting glucose, triacylglycerols and adiposity. Genetic analyses were performed to determine whether these findings might be confounded by obesity or insulin resistance, which they were not.
UK Biobank has addressed some of the limitations of older cohorts by undertaking deep phenotyping at an unprecedented scale, with MRI scans being performed to determine tissue composition, serological samples collected for GWAS, metabolomic and telomere analyses, and validated health outcomes obtained through record linkage. A recent public–private partnership contributed a further UK£200m to undertake whole-genome sequencing of 500,000 UK Biobank participants and pilot work is underway to explore the use of proteomics assays.
In Europe, the Innovative Medicines Initiative (IMI), a partnership between the European Commission, top academic institutions, the European Federation of Pharmaceutical Industries and Associations (EFPIA) and other partners, has invested more than €230m in projects seeking to discover biomarkers that might lead to novel diabetes drug targets, enhance monitoring and/or aid the design of diabetes drug trials. Of these, one IMI consortium (Diabetes Research on Patient Stratification [DIRECT]) established new prospective cohort studies enrolling ~3000 participants at risk of or with newly diagnosed diabetes [30, 31]. The project’s primary objective was to discover biomarkers for glycaemic deterioration before and after the onset of type 2 diabetes and included extensive deep phenotyping at baseline and throughout follow-up. Several other IMI diabetes projects have relied predominantly on assimilating, assaying and mining data from existing epidemiological cohorts for the discovery of diabetes-relevant biomarkers (see Table 2). In the US, the Accelerating Medicines Partnership has genotyped DNA from multiple diabetes case−control studies and assimilated these and other genetic and phenotypic summary data to provide public-access genomics resources (e.g. www.type2diabetesgenetics.org) [32]. In Finland, the FinnGen project [33] has brought together universities, hospitals, biobanks and pharmaceutical companies to study the genetic bases of common and rare diseases, with a focus on biomedical innovation and drug development. In Sweden, academic institutions, hospitals, government and charitable trusts have partnered to research and deliver genomic medicines through Genome Medicine Sweden (https://genomicmedicine.se/en/) [34]. Elsewhere, the UK Government established a company (Genomics England) to deliver genomics medicine to the population of England, with several highly ambitious projects, including the 100,000 Genomes project [35], which is primarily focused on cancers and rare diseases, but which will also include ~8000 families with rare inherited metabolic and endocrine diseases.
Epidemiological cohorts are sometimes used to provide sampling frames from which participants with specific phenotypic or genetic characteristics are recalled for experimental studies or complex in vivo measurements. Recall-by-genotype studies are especially appealing, as the feature upon which participants are recalled (genotype) is not subject to change and this paradigm can be much more powerful for the assessment of gene–treatment interactions than conventional trials [36]. METSIM is a prospective cohort study of Finnish men. In a recent recall-by-genotype study (n = 45) nested within METSIM [37], p.Pro50Thr AKT2 variant carriers and common allele homozygous controls were recalled to investigate the effects of the p.Pro50Thr AKT2 variant on insulin-stimulated glucose uptake. In this study, carriers of the risk allele showed reductions in glucose uptake (39.4%) and rate of endogenous glucose production (55.6%) after insulin stimulation compared with non-carriers. Glucose uptake was reduced primarily in musculoskeletal tissue.
Analytical challenges and emerging solutions
The analysis of dense multiomics datasets has proven formidable. To address some of the computational challenges, machine learning methods have been applied to determine hidden structures that are informative of disease aetiology or prognosis [38, 39]. Emphasis has been placed on the reclassification of the diagnosis of type 2 diabetes into subtypes. The principle of subclassifying diabetes using genetics and applying this knowledge to guide therapeutic decision making has proof of principle in the monogenic form of diabetes called MODY, which is characterised by defects in the development of the pancreatic islet cells and insulin secretion. The effective stratification of polygenic diabetes (type 1, type 2 and gestational diabetes), while highly appealing, is more challenging though, as complex diabetes manifests through defects in multiple organs, tissues and pathways [40] and is influenced by a wide range of environmental risk factors [41].
The stratification approaches reported to date for polygenic diabetes have used clinical phenotypes (e.g. BMI, C-peptide or HbA1c) [42], continuous glucose monitor-derived data [43] or genotypes [44,45,46] to stratify diabetes into aetiological subclasses. One of the earliest attempts to do this derived three diabetes subtypes by clustering data from electronic medical records and regressed genotype array data against each subtype to provide sets of SNPs from which pathophysiological inferences were made [44]. This approach is prone to type 1 error, owing to the large number of parallel hypothesis tests performed, the liberal significance threshold employed when selecting SNPs and the manner in which biological function was assigned to SNP sets (which may be prone to bias owing to the type of data available at that time). By contrast, the more recent studies using SNP clustering approaches [45, 46] are less prone to bias or type 1 error, as a very conservative p value threshold is used when selecting SNPs and the pathogenicity of variants is determined through very large and well-phenotyped independent datasets. Key barriers to the clinical translation of these approaches is that most use probabilistic soft clustering methods, which do not classify most individuals into discrete subtypes of diabetes, but instead assign one or more probabilities linking the individual to one or more subtypes of diabetes.
Udler et al derived process-specific clusters using enhancer enrichment from multi-cell epigenomic data [46], which were used to inform the design of polygenic risk scores (PRS), where higher scores were associated with relevant clinical outcomes (e.g. hypertension, coronary artery disease and stroke). These process-specific clusters characterised: (1) elevated beta cell function, (2) diminished beta cell function, (3) insulin resistance, (4) lipodystrophy-like adipose distribution and (5) disrupted liver lipid metabolism. Mahajan et al described similar clusters [45], but did not proceed to link these with clinical traits through participant-level association analyses. Thus, using these approaches to re-diagnose an individual with a new subtype of diabetes in a clinically meaningful and actionable manner is challenging.
Overall, these studies have provided intriguing insights into the aetiology of diabetes and helped to elucidate the factors that drive disease progression. However, many of these clustering methods do not assign most individuals to distinct clusters or risk misclassifying people to incorrect diagnostic categories (because most people are not defined by a distinct subtype of diabetes). Those methods that do not seek to assign individuals to distinct clusters focus on assigning probabilities of belonging to one or more clusters, which may be difficult to utilise in current clinical settings and may be less powerful than algorithms using continuous data [47].
Richardson et al [48] have categorised the existing omics integration approaches into vertical and horizontal methods. Vertical integration can be viewed as the combination of multiple ‘layers’ of data usually derived from the same individual. By contrast, horizontal data integration incorporates the same type of data derived from separate cohorts. Ritchie et al [49] describe multi-staged analyses, where associations are tested within data types (i.e. SNP datasets), filtered and then tested against traits, with the limitation of assuming linearity; meta-dimensional analysis simultaneously integrates various data types into a single model.
In a step towards clinical translation of omics data, a recent analysis assigned the participants whole-genome sequences pathogenicity scores according to the American College of Medical Genetics guidelines, revealing that one in every six participants carried at least one pathogenic variant [50]. Clinical biochemistry, metabolomic and digital imaging data (from MRI, CT, ECG, echocardiography, continuous glucose monitoring), as well as information from the participant’s medical records and family history were subsequently combined to derive a set of clinically relevant phenotypes relating mainly to cardiac and endocrine disorders (including type 2 and syndromic forms of diabetes). Associations were then tested between pathogenic variants and these clinical phenotypes, revealing that one in nine participants carried pathogenic variants that mapped to relevant clinical traits [50]. These findings imply that the appropriate use of deep-phenotyping data may enhance the ability to discriminate between high- and low-risk individuals with conventional risk factors and/or disease characteristics.
Summary and conclusions
The major expansion of accessible omics data in large epidemiological cohorts provides unprecedented opportunities for diabetes research and practice. Breakthroughs in knowledge will require training in analytical methods to keep pace with data generation; with very large and complex datasets, tasks that were once considered simple, such as data handling and quality control, now often require specialist training. The analyses that follow, possibly focusing on casual inference, gene–environment interactions, pharmacogenomics or functional annotation, will require other types of specialist knowledge. Many of these analyses will make use of external datasets that help establish biologically meaningful connections between molecular phenotypes, which requires specialist knowledge to access, integrate and interpret this information. Thus, appropriate training in specialist analytical tasks will be increasingly important for the next generation of epidemiologists. Importantly though, this should be balanced against the need for education in the core tenets of epidemiology, so that conclusions drawn from complex analyses are accurate and meaningful.
Abbreviations
- EPIC:
-
European Prospective Investigation into Cancer and Nutrition
- FDA:
-
Food and Drug Administration
- GWAS:
-
Genome-wide association studies
- IMI:
-
Innovative Medicines Initiative
- MR:
-
Mendelian randomisation
References
Seifert WK, Teeter RM (1969) Preparative thin-layer chromatography and high-resolution mass spectrometry of crude oil carboxylic acids. Anal Chem 41(6):786–795
Karpetsky TP, Humphrey RL, Levy CC (1977) Influence of renal insufficiency on levels of serum ribonuclease in patients with multiple myeloma. J Natl Cancer Inst 58(4):875–880
Biomarkers Definitions Working Group, Atkinson AJ Jr, Colburn WA et al (2001) Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther 69(3):89–95
US Food and Drug administration (FDA) (2018) Context of use. Available from https://www.fda.gov/drugs/cder-biomarker-qualification-program/context-use. Accessed 12 Jan 2020
FDA-NIH Biomarker Working Group (2016) BEST (Biomarkers, EndpointS, and other Tools) Resource. Food and Drug Administration (US), Silver Spring; National Institutes of Health (US), Bethesda
Biomarkers Consortium Evidentiary Standards Writing Group (2016) Framework for defining evidentiary criteria for biomarker qualification. Available from https://fnih.org/sites/default/files/final/pdf/EvidentiaryCriteriaFrameworkFinalVersionOct202016.pdf. Accessed 12 Jan 2020
Evangelou E, Ioannidis JP (2013) Meta-analysis methods for genome-wide association studies and beyond. Nat Rev Genet 14(6):379–389. https://doi.org/10.1038/nrg3472
Flannick J (2019) The contribution of low-frequency and rare coding variation to susceptibility to type 2 diabetes. Curr Diab Rep 19(5):25
Kowalski MH, Qian H, Hou Z et al (2019) Use of >100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations. PLoS Genet 15(12):e1008500
McCarthy S, Das S, Kretzschmar W et al (2016) A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 48(10):1279–1283. https://doi.org/10.1038/ng.3643
Moltke I, Grarup N, Jorgensen ME et al (2014) A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature 512(7513):190–193. https://doi.org/10.1038/nature13425
Manning M, Hudgins L (2010) Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet Med 12(11):742. https://doi.org/10.1097/GIM.0b013e3181f8baad
Tabák AG, Jokela M, Akbaraly TN, Brunner EJ, Kivimäki M, Witte DR (2009) Trajectories of glycaemia, insulin sensitivity, and insulin secretion before diagnosis of type 2 diabetes: an analysis from the Whitehall II study. Lancet 373(9682):2215–2221. https://doi.org/10.1016/S0140-6736(09)60619-X
Bradford Hill A (2015) The environment and disease: association or causation? J R Soc Med 108(1):32–37. https://doi.org/10.1177/0141076814562718
Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G (2008) Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 27(8):1133–1163. https://doi.org/10.1002/sim.3034
Laakso M (2019) Biomarkers for type 2 diabetes. Mol Metab 27s:S139–s146. https://doi.org/10.1016/j.molmet.2019.06.016
Lotta LA, Scott RA, Sharp SJ et al (2016) Genetic predisposition to an impaired metabolism of the branched-chain amino acids and risk of type 2 diabetes: a Mendelian randomisation analysis. PLoS Med 13(11):e1002179. https://doi.org/10.1371/journal.pmed.1002179
Ye Z, Sharp SJ, Burgess S et al (2015) Association between circulating 25-hydroxyvitamin D and incident type 2 diabetes: a mendelian randomisation study. Lancet Diabetes Endocrinol 3(1):35–42. https://doi.org/10.1016/S2213-8587(14)70184-6
Jorde R, Schirmer H, Wilsgaard T et al (2012) Polymorphisms related to the serum 25-hydroxyvitamin D level and risk of myocardial infarction, diabetes, cancer and mortality. The Tromsø Study. PLoS One 7(5). https://doi.org/10.1161/CIRCULATIONAHA.112.119693
Afzal S, Brøndum-Jacobsen P, Bojesen SE, Nordestgaard BG (2014) Vitamin D concentration, obesity, and risk of diabetes: a mendelian randomisation study. Lancet Diabetes Endocrinol 2(4):298–306. https://doi.org/10.1016/S2213-8587(13)70200-6
Lu L, Bennett DA, Millwood IY et al (2018) Association of vitamin D with risk of type 2 diabetes: a Mendelian randomisation study in European and Chinese adults. PLoS Med 15(5):e1002566. https://doi.org/10.1371/journal.pmed.1002566
Abbasi A (2015) Mendelian randomization studies of biomarkers and type 2 diabetes. Endocr Connect 4(4):249–260. https://doi.org/10.1530/EC-15-0087
Abbasi A, Sahlqvist A-S, Lotta L et al (2016) A systematic review of biomarkers and risk of incident type 2 diabetes: an overview of epidemiological, prediction and aetiological research literature. PLoS One 11(10):e0163721. https://doi.org/10.1371/journal.pone.0163721
Davey Smith G, Ebrahim S (2003) ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 32(1):1–22. https://doi.org/10.1093/ije/dyg070
Wootton RE, Lawn RB, Millard LA et al (2018) Evaluation of the causal effects between subjective wellbeing and cardiometabolic health: Mendelian randomisation study. BMJ 362:k3788. https://doi.org/10.1136/bmj.k3788
Loft S, Poulsen HE (1996) Cancer risk and oxidative DNA damage in man. J Mol Med 74(6):297–312. https://doi.org/10.1007/BF00207507
Eastwood SV, Mathur R, Atkinson M et al (2016) Algorithms for the capture and adjudication of prevalent and incident diabetes in UK Biobank. PLoS One 11(9):e0162388. https://doi.org/10.1371/journal.pone.0162388
Chen R, Mias GI, Li-Pook-Than J et al (2012) Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 148(6):1293–1307. https://doi.org/10.1016/j.cell.2012.02.009
Imamura F, Sharp SJ, Koulman A et al (2017) A combination of plasma phospholipid fatty acids and its association with incidence of type 2 diabetes: the EPIC-InterAct case-cohort study. PLoS Med 14(10):e1002409. https://doi.org/10.1371/journal.pmed.1002409
Koivula RW, Heggie A, Barnett A et al (2014) Discovery of biomarkers for glycaemic deterioration before and after the onset of type 2 diabetes: rationale and design of the epidemiological studies within the IMI DIRECT Consortium. Diabetologia 57(6):1132–1142. https://doi.org/10.1007/s00125-014-3216-x
Rauh SP, Heymans MW, Koopman AD et al (2017) Predicting glycated hemoglobin levels in the non-diabetic general population: development and validation of the DIRECT-DETECT prediction model-a DIRECT study. PLoS One 12(2):e0171816. https://doi.org/10.1371/journal.pone.0171816
Accelerating Medicines Partnership (AMP): Type 2 Diabetes (2020) Type 2 Diabetes Knowledge Portal. Available from www.type2diabetesgenetics.org/. Accessed 12 Jan 2020
The FinnGen project (2019) The FinnGen study. Available from www.finngen.fi/en. Accessed 12 Jan 2020
The Genomic Medicine Sweden (GMS) initiative (2019) Genomic Medicine Sweden. Available from https://genomicmedicine.se/en/. Accessed 18 Dec 2019
Genomics England (2020) 100,000 Genomes Project. Available from https://www.genomicsengland.co.uk/about-genomics-england/the-100000-genomes-project. Accessed 18 Dec 2019
Franks PW, Timpson NJ (2018) Genotype-based recall studies in complex cardiometabolic traits. Circ Genom Precis Med 11(8):e001947
Latva-Rasku A, Honka M-J, Stančáková A et al (2018) A partial loss-of-function variant in AKT2 is associated with reduced insulin-mediated glucose uptake in multiple insulin-sensitive tissues: a genotype-based callback positron emission tomography study. Diabetes 67(2):334–342. https://doi.org/10.2337/db17-1142
Pare G, Mao S, Deng WQ (2017) A machine-learning heuristic to improve gene score prediction of polygenic traits. Sci Rep 7(1):1–11
Dworzynski P, Aasbrenn M, Rostgaard K et al (2020) Nationwide prediction of type 2 diabetes comorbidities. Sci Rep 10(1):1–13
McCarthy MI (2017) Painting a new picture of personalised medicine for diabetes. Diabetologia 60(5):793–799. https://doi.org/10.1007/s00125-017-4210-x
Mutie PM, Giordano GN, Franks PW (2017) Lifestyle precision medicine: the next generation in type 2 diabetes prevention? BMC Med 15(1):171
Ahlqvist E, Storm P, Käräjämäki A et al (2018) Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol 6(5):361–369. https://doi.org/10.1016/S2213-8587(18)30051-2
Hall H, Perelman D, Breschi A et al (2018) Glucotypes reveal new patterns of glucose dysregulation. PLoS Biol 16(7):e2005143. https://doi.org/10.1371/journal.pbio.2005143
Li L, Cheng W-Y, Glicksberg BS et al (2015) Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Sci Transl Med 7(311):311ra174–311ra174. https://doi.org/10.1126/scitranslmed.aaa9364
Mahajan A, Wessel J, Willems SM et al (2018) Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat Genet 50(4):559–571. https://doi.org/10.1038/s41588-018-0084-1
Udler MS, Kim J, von Grotthuss M et al (2018) Type 2 diabetes genetic loci informed by multi-trait associations point to disease mechanisms and subtypes: a soft clustering analysis. PLoS Med 15(9):e1002654. https://doi.org/10.1371/journal.pmed.1002654
Dennis JM, Shields BM, Henley WE, Jones AG, Hattersley AT (2019) Disease progression and treatment response in data-driven subgroups of type 2 diabetes compared with models based on simple clinical features: an analysis using clinical trial data. Lancet Diabetes Endocrinol 7(6):442–451. https://doi.org/10.1016/S2213-8587(19)30087-7
Richardson S, Tseng GC, Sun W (2016) Statistical methods in integrative genomics. Annu Rev Stat Appl 3(1):181–209. https://doi.org/10.1146/annurev-statistics-041715-033506
Ritchie MD, Holzinger ER, Li R, Pendergrass SA, Kim D (2015) Methods of integrating data to uncover genotype–phenotype interactions. Nat Rev Genet 16(2):85–97. https://doi.org/10.1038/nrg3868
Hou Y-CC, Yu H-C, Martin R et al (2020) Precision medicine integrating whole-genome sequencing, comprehensive metabolomics, and advanced imaging. Proc Natl Acad Sci 117(6):3053–3062. https://doi.org/10.1073/pnas.1909378117
Yadav SP (2007) The wholeness in suffix-omics,-omes, and the word om. J Biomol Tech 18(5):277
Cook A (1977) The William Bateson papers. The Mendel newsletter; archival resources for the history of genetics & allied sciences. 14:1. https://doi.org/10.5694/j.1326-5377.1977.tb99336.x
Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM (1998) Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol 5(10):R245–R249. https://doi.org/10.1016/S1074-5521(98)90108-9
Peregrin T (2001) The new frontier of nutrition science: nutrigenomics. J Acad Nutr Diet 101(11):1306
Wilkins MR, Sanchez J-C, Gooley AA et al (1996) Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it. Biotechnol Genet Eng Rev 13(1):19–50. https://doi.org/10.1080/02648725.1996.10647923
Nicholson JK, Lindon JC, Holmes E (1999) ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 29(11):1181–1189
Waddington CH (2012) The epigenotype. Int J Epidemiol 41(1):10–13. https://doi.org/10.1093/ije/dyr184
Callinan PA, Feinberg AP (2006) The emerging science of epigenomics. Hum Mol Genet 15(suppl_1):R95–R101. https://doi.org/10.1093/hmg/ddl095
Turnbull JE, Field RA (2007) Emerging glycomics technologies. Nat Chem Biol 3(2):74–77. https://doi.org/10.1038/nchembio0207-74
Han X, Gross RW (2003) Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry a bridge to lipidomics. J Lipid Res 44(6):1071–1079. https://doi.org/10.1194/jlr.R300004-JLR200
Piétu G, Mariage-Samson R, Fayein N-A et al (1999) The Genexpress IMAGE knowledge base of the human brain transcriptome: a prototype integrated resource for functional and computational genomics. Genome Res 9(2):195–209
Fagerholm E, Ahlqvist E, Forsblom C et al (2012) SNP in the genome-wide association study hotspot on chromosome 9p21 confers susceptibility to diabetic nephropathy in type 1 diabetes. Diabetologia 55(9):2386–2393. https://doi.org/10.1007/s00125-012-2587-0
Heinzel A, Kammer M, Mayer G et al (2018) Validation of plasma biomarker candidates for the prediction of eGFR decline in patients with type 2 diabetes. Diabetes Care 41(9):1947–1954. https://doi.org/10.2337/dc18-0532
Koivula RW, Forgie IM, Kurbasic A et al (2019) Discovery of biomarkers for glycaemic deterioration before and after the onset of type 2 diabetes: descriptive characteristics of the epidemiological studies within the IMI DIRECT Consortium. Diabetologia 62(9):1601–1615. https://doi.org/10.1007/s00125-019-4906-1
Authors’ relationships and activities
PWF has received research funding from Boehringer Ingelheim, Eli Lilly, Janssen, Novo Nordisk A/S, Sanofi Aventis and Servier, received consulting fees from Eli Lilly, Novo Nordisk and Zoe Global Ltd and has stock options in Zoe Global Ltd.
Funding
Open access funding provided by Lund University. PWF and HPM are funded by: European Research Council (no. CoG-2015_681742_NASCENT); Swedish Research Council; Novo Nordisk Foundation; European Diabetes Research Foundation; Swedish Heart Lung Foundation; Innovative Medicines Initiative of the European Union (no. 115317 – DIRECT and no. 115881 – RHAPSODY; no. 875534 – SOPHIA); Swedish Foundation for Strategy Research (no. IRC15-0067).
Author information
Authors and Affiliations
Contributions
Both authors conceived the ideas described herein, reviewed relevant literature and co-wrote the manuscript.
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Figure slide
(PPTX 162 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Franks, P.W., Pomares-Millan, H. Next-generation epidemiology: the role of high-resolution molecular phenotyping in diabetes research. Diabetologia 63, 2521–2532 (2020). https://doi.org/10.1007/s00125-020-05246-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-020-05246-w