Abstract
Aims/hypothesis
Obesity and insulin resistance may be associated with altered expression and secretion of adipokines. Physical activity can markedly improve insulin sensitivity, but the association with adipokines remains largely unknown. In this study, we examined the effects of physical activity on the subcutaneous white adipose tissue (scWAT) secretome and its relationship to insulin sensitivity.
Methods
As reported previously, we enrolled 26 sedentary, middle-aged men (13 dysglycaemic and overweight; 13 normoglycaemic and of healthy weight) into a 12 week, supervised, intensive physical exercise intervention that included two endurance and two resistance sessions each week. Insulin sensitivity was measured as the glucose infusion rate from a euglycaemic–hyperinsulinaemic clamp. In our previous study, we measured maximum oxygen uptake, upper- and lower-body strength and a range of circulating biomarkers, and quantified adipose tissue depots using MRI and magnetic resonance spectroscopy. We have now performed global mRNA sequencing, microarrays and RT-PCR of scWAT and skeletal muscle biopsies, and quantified selected plasma adipokines by ELISA.
Results
Insulin sensitivity increased similarly in both dysglycaemic (45%) and normoglycaemic (38%) men after 12 weeks of exercise, as reported previously. mRNA sequencing of scWAT revealed 90 transcripts that responded to exercise in dysglycaemic men, whereas only marginal changes were observed in normoglycaemic men. These results were validated using microarrays and RT-PCR. A total of 62 out of 90 transcripts encoded secreted proteins. Overall, 17 transcripts were upregulated and 73 transcripts were downregulated. Downregulated transcripts included several macrophage markers, and were associated with inflammatory and immune-related pathways. Levels of these immune-related transcripts were enhanced in dysglycaemic men vs normoglycaemic men at baseline, but were normalised after the exercise intervention. Principal component and correlation analyses revealed inverse correlations between levels of these immune-related transcripts and insulin sensitivity at baseline, after the intervention, and for the change between baseline and after the intervention. In addition, levels of these transcripts at baseline could predict exercise-induced improvements in insulin sensitivity. Adipokine levels in scWAT (but not in skeletal muscle) were significantly correlated with corresponding plasma adipokine concentrations, as exemplified by leptin, high-molecular-weight adiponectin and secreted frizzled-related protein 4 (SFRP4). SFRP4 mRNA was the most exercise-responsive transcript in scWAT from dysglycaemic men, and plasma SFRP4 concentrations were reduced in dysglycaemic men, but not in normoglycaemic men, after 12 weeks of exercise.
Conclusions/interpretation
This study indicates that scWAT may be an important mediator of exercise-induced improvements in insulin sensitivity, especially in overweight dysglycaemic individuals at increased risk of developing type 2 diabetes.
Similar content being viewed by others

Introduction
Adipose tissue is an active endocrine organ that expresses and secretes multiple metabolically active factors such as leptin, adiponectin, IL-6 and TNF-α [1,2,3]. These secreted factors are involved in metabolic and inflammatory processes and may act in a paracrine or endocrine way, altering metabolism in the liver, pancreas, skeletal muscle and central nervous system [3, 4]. Dysregulation of these signal molecules is closely related to adipocyte hypertrophy and insulin resistance [5,6,7], and has been characterised in several studies [3, 8,9,10].
Insulin resistance is a hallmark of type 2 diabetes mellitus and is closely linked to lifestyle variables such as diet and physical activity [11,12,13,14]. Physical activity can substantially increase insulin sensitivity, and exercise-induced alterations in subcutaneous white adipose tissue (scWAT) may affect whole-body metabolic health [4, 15, 16]. Mediators of these effects may involve extensive adaptations in adipokine expression [4, 16]. Numerous studies have focused on the effect of different types of exercise on circulating levels of adipose tissue-derived factors [3, 17,18,19]. However, the main body of literature on adipokines and exercise is limited to plasma analyses of one or a few targets and the effects of acute exercise [3, 19,20,21,22] confounded by weight loss [23] or focused on only visceral adipose tissue [3, 17, 18]. Few studies have addressed long-term exercise-induced alterations in scWAT expression of such factors [22, 24], and these studies have only been performed in insulin-sensitive women [22, 24]. In addition, whereas skeletal muscle has been extensively studied as regards physical exercise [25], significantly less focus has been placed on adipose tissue in this aspect.
In the present study, we investigated the effect of long-term physical exercise on scWAT transcript levels and potential links to insulin sensitivity. We performed global mRNA sequencing on biopsies, together with ELISA measurements of selected plasma adipokines, and quantified insulin sensitivity using a euglycaemic–hyperinsulinaemic clamp. We hypothesised that 12 weeks of combined endurance and strength training would promote distinct alterations in the scWAT transcriptome among overweight men with dysglycaemia and men of healthy weight with normoglycaemia, and that these changes would correlate with insulin sensitivity.
Methods
Participants and experimental methods standardisation
The MyoGlu study was a controlled clinical exercise intervention trial (ClinicalTrials.gov registration no. NCT01803568) in 26 sedentary white (<1 exercise session/week) men aged 40–65 years of Scandinavian origin performed in 2011–2012 in Oslo, Norway (Fig. 1) [26]. All participants gave written informed consent, and the study was approved by the Regional Committee for Medical and Health Research Ethics North (Tromsø, Norway; ref. no. 2011/882) and adhered to the Declaration of Helsinki. The study was designed to in-depth phenotypically characterise the effects of an intensive exercise intervention across the glucometabolic spectrum.
Study design. (a) A total of 26 sedentary men (13 of healthy weight with normoglycaemia [control] and 13 overweight with dysglycaemia) were recruited into two groups. 1No participants were excluded because of lack of available time; a very small number were excluded because they did not meet the age criterion, ~70% were excluded because of medication or because of family members with diabetes (controls); ~30% were excluded because they had been too active in the past year. 2OGTT response did not match phenotype, e.g. an obese person had a normal OGTT response. 3One participant was diagnosed with acute coronary syndrome. 4Due to illness. (b) The participants donated tissue samples and underwent several tests at baseline, including a euglycaemic–hyperinsulinaemic clamp, before undertaking 12 weeks of an intensive physical exercise intervention followed by re-testing. Three days passed between the last bout of exercise and the euglycaemic–hyperinsulinaemic clamp after the intervention. (c) The intervention consisted of four exercise sessions each week
We included 13 dysglycaemic, overweight men (fasting glucose ≥5.6 mmol/l, 2 h OGTT ≥7.8 mmol/l or insulin resistance [HOMA-IR >2.0]; BMI 26.8–32.5 kg/m2) and 13 normoglycaemic men of healthy weight (fasting glucose <5.6 mmol/l and 2 h OGTT <7.8 mmol/l and without a family history of diabetes; BMI 20.9–26.7 kg/m2). Exclusion criteria included smoking, a family history of diabetes (for control participants only), known hypertension, liver or kidney disease, chronic inflammatory diseases or use of a medication expected to affect glucose metabolism (e.g. lipid-lowering or anti-hypertensive medications, acetylsalicylic acid, corticosteroids).
Before the intervention, the participants refrained from physical exercise and alcohol for 2 days before testing (Fig. 1). The last session of the 12 week intervention consisted of an endurance session with intervals performed at 85% of maximum heart rate 3 days prior to repeated testing (Fig. 1). Maximal oxygen uptake (\( \dot{V} \)O2max) tests and maximum strength tests were performed several days (>3 days) before the euglycaemic–hyperinsulinaemic clamp and tissue sampling (Fig. 1). Tests were performed under similar conditions on separate days both before and after the intervention, with some exceptions for MRI/magnetic resonance spectroscopy (MRS) due to scanner availability.
Diet
The habitual diet of each participant was recorded using a validated food frequency questionnaire [27, 28]. Calculations were performed using the food database AE-10 and the KBS v.7.1 (Kostberegningssystem, Oslo, Norway) food and nutrients calculation system. Alcohol intake was not allowed to exceed 2 units per day. During testing at baseline and after 12 weeks of exercise, the participants consumed a standardised meal after an overnight fast. A carbohydrate-rich meal including bread, apple juice, cheese and jam was adjusted depending on individual energy requirements and provided 23% of the estimated total daily energy expenditure 90–120 min prior to the test. Tests were typically performed in the morning; the standardised meal was the only intake after the overnight fast. Water could be consumed freely.
Exercise intervention
Strength and endurance exercise
The participants performed 4 h of intensive exercise each week for 12 weeks under professional supervision. Two whole-body strength training sessions and two spinning bike interval sessions lasted 1 h each (Fig. 1). The 12 week intervention included linear progression in workload for both strength and endurance exercises. For strength exercises, during weeks 1–3 a load that could be lifted a maximum of 12 times (i.e. 12 repetition maximum) was used, which progressed to ten repetition maximum in weeks 4–8 and to eight repetition maximum in weeks 9–12. Abdominal crunches and back extensions were performed with 12–20 repetitions for the whole intervention period. For endurance exercise, during week 1, three intervals were performed during endurance session one (intervals at >90% of maximum heart rate) and six intervals during endurance session two (intervals at 85% of maximum heart rate) (Fig. 1). For weeks 2–5, four intervals were performed during session one and seven intervals during session two. For weeks 6–12, five intervals were performed during session one and ten intervals during session two.
Physical fitness and insulin sensitivity
\( \dot{V}{\mathrm{O}}_{2\mathrm{max}} \) tests were performed after a standardised warm-up at a workload similar to the final load of an incremental test in which the relationship between work (watt) and oxygen uptake was established. Participants cycled for 1 min followed by a 15 W increased workload every 30 s until exhaustion. Test success was based on oxygen consumption increased by <0.5 ml kg−1 min−1 over a 30 W increase in workload, respiratory exchange ratio values >1.10 and blood lactate >7.0 mmol/l.
Euglycaemic–hyperinsulinaemic clamp
Euglycaemic–hyperinsulinaemic clamps were performed after an overnight fast. A fixed dose of insulin 40 mU m–2 min–1 was infused, and glucose (200 mg/ml) was injected to maintain euglycaemia (5.0 mmol/l) for 150 min [29]. Insulin sensitivity is reported as the glucose infusion rate (GIR) during the last 30 min relative to body weight. The whole blood glucose concentration was measured using a glucose oxidase method (YSI 2300, YSI, Yellow Springs, OH, USA) and the plasma glucose concentration was calculated as whole blood glucose ×1.119.
MRI/MRS
MRI methods were used to quantify fat mass [26]. The ankle-to-neck MRI protocol included a 3D Dixon acquisition providing water and lipid quantification. Data were then analysed using the nordicICE software package (NordicNeuroLab, Bergen, Norway) and the jMRUI v.5.2 [30] software package.
Tissue sampling
As described previously [26], we obtained scWAT, skeletal muscle biopsies and blood samples before and after the 12 week exercise intervention (Fig. 1). Biopsies were obtained from the periumbilical subcutaneous tissue and from the vastus lateralis muscle. After sterilisation, a lidocaine-based local anaesthetic was injected into the skin and subcutis prior to both skeletal muscle and scWAT biopsies [26]. Biopsies were dissected on a cold aluminium plate to remove blood and other materials before freezing.
Transcriptomics
Biopsies were frozen in liquid nitrogen, crushed to powder with a pestle in a liquid nitrogen-cooled mortar, transferred into 1 ml QIAzol Lysis Reagent (Qiagen, Hilden, Germany) and homogenised using TissueRuptor (Qiagen) at full speed for 15 s, twice. Total RNA was isolated from the homogenate using the miRNeasy Mini Kit (Qiagen). RNA integrity and concentration were determined using Agilent RNA 6000 Nano Chips on a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). RNA was converted to cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster, CA, USA). The cDNA reaction mixture was diluted in water and a cDNA equivalent of 25 ng RNA was used for each sample.
TaqMan real-time quantitative RT-PCR
Quantitative real-time PCR was performed with reagents and instruments from Applied Biosystems in the 96-well format using a 7900HT Fast instrument and SDS 2.3 software (Applied Biosystems). Predeveloped primers and probe sets (TaqMan assays, Applied Biosystems) were used to analyse mRNA levels of SFRP4 (Hs00180066_m1), LEP (Hs00174877_m1), TNFRSF11B (Hs00900358_m1), IL6 (Hs00985639_m1) and ADIPOQ (Hs00605917_m1). Relative target mRNA levels were calculated as \( {2}^{-\Delta {\mathrm{C}}_{\mathrm{t}}} \) and normalised to B2M (Hs00984230_m1).
Microarrays
Purified RNA was labelled using the Affymetrix WT PLUS reagent kit (Affymetrix, Santa Clara, CA, USA) and hybridised to an Affymetrix Human Gene 1.1 ST array plate. Hybridisation, washing and scanning were carried out on the Affymetrix GeneTitan platform according to the manufacturer’s instructions. Arrays were analysed using the R package Oligo v.1.46 [31] following standard procedures for quality checks and calculation of normalised expression values.
High-throughput mRNA sequencing
All muscle and scWAT samples were deep-sequenced using the Illumina HiSeq 2000 system (San Diego, CA, USA) with multiplex at the Norwegian Sequencing Centre, University of Oslo. Illumina HiSeq RTA (real-time analysis) v1.17.21.3 was used. Reads passing Illumina’s recommended parameters were demultiplexed using CASAVA v1.8.2 (Illumina). For pre-alignment quality checks, we used the software FastQC v0.10.1 (Illumina). The mean library size was approximately 44 million unstranded 51 bp single-ended reads for muscle tissue and approximately 52 million for scWAT, with no differences between groups or time points. No batch effects were present. cDNA sequenced reads alignment was performed using Tophat v2.0.8 [32], Samtools v0.1.18 [33] and Bowtie v2.1.0 [34] with default settings against the UCSC hg19 (University of California Santa Cruz, Santa Cruz, CA, USA) annotated transcriptome and genome dated 14 May 2013. Post-alignment quality controls were performed using the Integrative Genome Viewer v2.3 (Illumina) and BED tools v2.19.1 [35]. Reads were counted using the intersection strict mode in HTSeq v0.6.1 [36].
Differential transcript expression using mRNA sequencing
The edgeR v3.4.2 [37], DESeq2 v1.4.5 [38], and Cuffdiff v2.1.1 [32] workflows were performed. Statistical significance was set at a false discovery rate of <15% for each approach, and then intersected to find coherent results from the three approaches. TaqMan real-time RT-PCR and microarrays were subsequently used to validate the results. Expression levels are presented as reads per kilobase of transcript per million mapped reads.
Pathway analysis of mRNA sequencing results
Pathway analysis was performed using MSigDB (The Molecular Signatures Database; http://software.broadinstitute.org/gsea/msigdb) KEGG pathways. Differentially expressed transcripts were tested for significant overlaps with these pathways using hypergeometric tests [39]. The Benjamini–Hochberg procedure was used to correct p values [40]. Upstream transcriptional regulators of the observed transcript changes were identified using Qiagen Upstream Regulator Analysis (Hilden, Germany).
Secretory proteins
We used the MetazSecKB (the metazoa [human & animal] protein subcelluar location, secretome and subcellular proteome database; http://bioinformatics.ysu.edu/secretomes/animal/index.php) knowledgebase to identify transcripts encoding secreted proteins. MetazSecKB identifies secretory proteins based on either curated evidence of secretion (annotated and reviewed in the UniProtKB/Swiss-Prot dataset) or computationally predicted secretory protein sequences, without containing transmembrane domains or endoplasmic reticulum retention signals, using several tools (SignalP4 [41], Phobius [42], TargetP [43] and WoLF PSORT [44]).
scWAT cell type transcript markers
We selected high-specificity markers of scWAT cell types (adipocytes, macrophages, leucocytes and progenitor cells) based on the study by Ehrlund et al [45], and markers of M1-like and M2-like macrophages from the study by Hill et al [46].
Plasma analyses
Plasma samples of secreted frizzled-related protein 4 (SFRP4) (catalogue no. SEF878Hu, Cloud-Clone, Houston, TX, USA), total and high-molecular-weight adiponectin (catalogue no. DRP300 and DHWAD0, R&D Systems, Minneapolis, MN, USA), IL-6 (catalogue no. HS600B, R&D Systems) and leptin (catalogue no. KAC2281, Invitrogen, Carlsbad, CA, USA) were measured in duplicate using ELISA according to the manufacturer’s protocol. Absorbance was determined using a microplate reader (Titertec Multiscan Plus; EFLAB, Helsinki, Finland), which was set to 450 or 490 nm depending on the specific protocol. Standard curves for all proteins were generated using a best-fit curve.
External datasets
We compared our results with those from two other independent studies in obese, insulin-resistant participants by using the Array Express (EMBL-EBI), and the E-GEOD-70529 [47] and E-GEOD-26637 [48] datasets, respectively. The R packages Oligo [31] and LIMMA [49] were used for analysis.
Statistics
Data were modelled using parametric or non-parametric methods, as appropriate and specified along with each analysis. p values were considered significant at α = 0.05. All data were analysed using R v3.3.3 [50]. We performed principal component analysis to enable the study of intercorrelated measures by producing linear combinations (principal components), where the first principal component is the linear sum of the measures that has the largest total variance [51].
Results
GIR increased by 45% and 38% in men with dysglycaemia and normoglycaemia, respectively (Table 1). Participant characteristics are presented in Table 1. At baseline, men with dysglycaemia had significantly more adipose tissue, lower \( \dot{V}{\mathrm{O}}_{2\mathrm{max}} \) and impaired glucose metabolism, and a non-significant tendency to higher plasma hsCRP levels (p < 0.06) vs control individuals. Body composition, physical fitness and glucose metabolism improved similarly for both groups in response to 12 weeks of exercise (Table 1), as reported previously [26].
scWAT transcriptomics response to 12 weeks of exercise
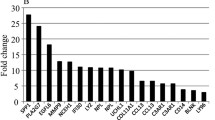
Intersected results from all three approaches to mRNA sequencing analyses revealed seven transcripts responding to 12 weeks of exercise among control participants (Fig. 2a,c), as compared with 90 transcripts in men with dysglycaemia (Fig. 2b,d). Three transcripts were upregulated and four transcripts were downregulated in control participants (Fig. 2a and Table 2), while 17 transcripts were upregulated and 73 transcripts were downregulated in men with dysglycaemia (Fig. 2b and Table 3). Evidence for encoding secretory proteins existed for five of seven transcripts for control participants (Table 2) and for 62 of 90 transcripts for men with dysglycaemia (Table 3).
Exercise-responsive transcripts in scWAT. We compared transcript levels in biopsies obtained from scWAT before and after the 12 week exercise intervention using three commonly applied workflows for mRNA sequencing data: DESeq2, edgeR and Cuffdiff. The results from these three approaches were intersected for both (a) normoglycaemic and (b) dysglycaemic men. Volcano plots show fold changes and q values for (c) normoglycaemic and (d) dysglycaemic men. On the x-axis, data points below and above zero indicate downregulation and upregulation, respectively, after the 12 week exercise intervention. On the y-axis, large values represent low q values, represented as the mean from all three workflows. The logarithmic transformations used in volcano plots are necessary for comprehensive and symmetrical representation of the large amount of data. Red coloured points indicate statistical significance, whereas black points signifies statistical insignificance within each group, respectively. q values: Benjamini–Hochberg corrected p values
For the 73 transcripts that were downregulated in men with dysglycaemia, absolute levels of most mRNAs were higher in men with dysglycaemia as compared with control participants at baseline (Fig. 3). The differences in absolute transcript levels between the two groups were attenuated after 12 weeks of exercise (Fig. 3).
Twelve weeks of exercise normalised enhanced transcript levels in scWAT from men with dysglycaemia. The figure shows median levels of the 73 transcripts that were downregulated in men with dysglycaemia after 12 weeks of the exercise intervention. Each row represents the median level across all participants in the specified group, and each column represents one gene. The scale bar below the heat map represents centred and scaled reads per kilobase of transcript per million mapped reads
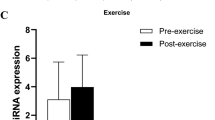
Our mRNA sequencing data were highly coherent across statistical approaches and across different technologies, including as RT-PCR (Table 4 and Fig. 4), but also cDNA microarrays from a subset of the samples (after 12 weeks of exercise, SFRP4 expression in control participants was reduced by 76.5% [p = 0.13; n = 4], whilst in men with dysglycaemia it was reduced by 55.3% [ p < 0.001 n =7]).
Validation of mRNA sequencing results. We analysed the transcript most downregulated in men with dysglycaemia after 12 weeks of exercise (SFRP4; see Table 3) using (a) mRNA sequencing (−28.9% [p>0.05] and −67.8 % [**p<0.01] in control individuals and men with dysglycaemia, respectively) and (b) real-time quantitative RT-PCR (−66.1% [p>0.05] and −112.4 % [**p<0.01] in control individuals and men with dysglycaemia, respectively). Paired t tests were used for comparisons. RPKM, reads per kilobase of transcript per million mapped reads
scWAT mRNA levels and insulin sensitivity
We applied principal component analyses to the 73 downregulated transcripts in men with dysglycaemia after 12 weeks of exercise, and correlated the first principal component to insulin sensitivity, measured as GIR (Fig. 5). The first principal component correlated with GIR at baseline (Fig. 5a) and after the 12 week intervention (Fig. 5b). We also observed significant correlations between changes in GIR in response to the 12 week intervention and changes in the levels of these transcripts (Fig. 5c), and baseline transcript levels predicted GIR change in response to the intervention (Fig. 5d). The transcripts exhibiting the most significant Spearman’s correlations with GIR are presented in Fig. 5e, and all transcript correlations with GIR are presented in the electronic supplementary material (ESM) Table 1.
Correlations between scWAT transcript levels and euglycaemic–hyperinsulinaemic clamp results. Principal component analysis was performed on the 73 transcripts downregulated after 12 weeks of exercise in men with dysglycaemia (see Fig. 3). The first principal component (PC1) correlated with GIR at (a) baseline and (b) after 12 weeks of exercise intervention, and (c) with change in GIR in response to 12 weeks of exercise intervention (principal component analysis was performed on change scores). (d) Transcript levels at baseline predicted the change in GIR in response to 12 weeks of exercise. Trend lines with standard error of estimates (SEEs; grey areas around the black regression lines) are presented. (e) The top ten most significant Spearman’s correlations between transcript levels and GIR (see also ESM Table 1). The results were similar using either Spearman’s or Pearson’s correlations. **p<0.01, ***p<0.001 for the correlation between the gene and GIR
scWAT pathway and upstream regulator analyses
Transcripts responding to 12 weeks of exercise in men with dysglycaemia (Table 3) overlapped with several immune-related pathways, such as leucocytes, transendothelial migration, toll-like receptor signalling, and B and T cell receptor signalling (Fig. 6a). These transcripts (Table 3) also correlated with plasma hsCRP levels (ESM Table 2), and might form a part of a cytokine signalling network regulated by the transcription factors SRY-box 9 (SOX9), transcriptional and immune response regulator (TCIM) and RAR-related orphan receptor C, isoform CRA_a (RORC) (Fig. 6b).
Pathways, cell types and mediation analyses. (a) Pathway-enrichment analysis of scWAT transcripts regulated after 12 weeks of exercise in dysglycaemic men, and (b) upstream regulatory network analysis. (c-h) Analyses of main cell populations in scWAT: (c) adipocytes, (d) macrophages, (e) leucocytes, (f) progenitor cells, (g) M1-like macrophages and (h) M2-like macrophages. Bar plots represent means ± SEM. *p<0.05, ***p<0.001 for between-group comparison (dysglycaemia vs control participants) at baseline using unpaired t tests. †p<0.05, ††p<0.01 for within-group comparison (after [light grey/light red] vs before [dark grey/dark red] the 12 week intervention) using paired t tests. (i, j) Mediation analyses between M2-like macrophage transcript levels and (i) plasma (P) adiponectin levels or (j) scWAT ADIPOQ transcript levels, and GIR. ‡ p<0.05, ‡‡p<0.01. DG, participants with dysglycaemia; RPKM, reads per kilobase of transcript per million mapped reads
scWAT cell populations
The 73 downregulated transcripts after 12 weeks of exercise in men with dysglycaemia were highly related to immune- and macrophage-related processes, based on gene-set enrichment analyses (Fig. 6a). We monitored the top 100 high-specificity transcript markers of human scWAT adipocytes, macrophages, leucocytes and progenitor cells, as according to Ehrlund et al [45]. Adipocyte-related transcript levels were lower and macrophage-related transcript levels were higher in men with dysglycaemia as compared with the control group (Fig. 6c,d). Macrophage- and leucocyte-related transcript levels decreased in men with dysglycaemia after 12 weeks of exercise intervention, whereas no changes were observed in the control participants (Fig. 6d,e). We also analysed high-specificity markers of human scWAT M1-like and M2-like macrophages, as according to Hill et al [46]. M2-like, but not M1-like, macrophage-related transcripts were more highly expressed in men with dysglycaemia as compared with control participants (Fig. 6g,h), and M2-like macrophage-related transcripts decreased in men with dysglycaemia after 12 weeks of the exercise intervention, whereas no change was observed in the control participants (Fig. 6h). M2-like macrophage transcript levels correlated negatively to GIR and plasma adiponectin levels (Fig. 6i) and scWAT ADIPOQ transcript levels (Fig. 6j) at baseline. However, the correlation between M2-like macrophage transcript levels and GIR disappeared after adjusting for adiponectin levels (Fig. 6i,j). Group differences and responses to 12 weeks of exercise for all of the top 100 transcript markers for each scWAT cell type are presented in ESM Tables 3–6.
Effects of weight loss and insulin
To explore potential mechanisms behind the dysglycaemia-specific effects of 12 weeks of exercise intervention on scWAT, we analysed the transcripts that changed in response to exercise in men with dysglycaemia only (Table 3). We investigated if the change in transcripts could be due to weight loss by comparing our exercise-changed transcripts to those changed in scWAT after weight loss in obese and insulin-resistant participants in the study of Magkos et al [47] (Fig. 7a). We also evaluated if the change in these transcripts might be due to hyperinsulinaemia by studying data from human scWAT before and after 3 h of insulin infusion in obese and insulin-resistant participants compared with lean and insulin-sensitive participants in the study by Soronen et al [48] (Fig. 7b,c).
Effects of weight loss and hyperinsulinaemia on subcutaneous adipose tissue transcript levels. (a) Effects of moderate and subsequent progressive weight loss from the study by Magkos et al [47]. Grey lines, p>0.05; red lines, p<0.05 (5, 10 or 15% weight loss vs baseline). (b) Effects of 3 h of insulin infusion (grey lines, p>0.05; red lines, p<0.05 3 h insulin infusion vs baseline), and (c) differences in response to insulin infusion between insulin-resistant (IR) and insulin-sensitive (IS) participants based on data from the study by Soronen et al [48]. The bar plot represents log2(fold change) ± 95% CIs for the response to insulin infusion in insulin-resistant vs insulin-sensitive participants. *p<0.05, **p<0.01, ***p<0.001
Thirteen transcripts were associated with genes changed in response to weight loss (Fig. 7a), and these exhibited the same direction of regulation as seen after 12 weeks of exercise intervention (Table 3). Eight transcripts in the study by Soronen et al [48] were associated with hyperinsulinaemia (Fig. 7b) and exhibited the opposite directions of regulation as seen after 12 weeks of exercise intervention (Table 3). Eight transcripts were differently regulated in response to insulin infusion between insulin-resistant and insulin-sensitive participants (Fig. 7c).
scWAT mRNA levels and plasma adipokine concentrations
LEP and ADIPOQ mRNA levels displayed significant correlations with plasma leptin and high-molecular-weight adiponectin concentrations in both groups of participants at baseline (Fig. 8). Similar results were also observed for IL-6 and total adiponectin, and for values after 12 weeks and for changes observed during the intervention (data not shown).
Correlations between transcript levels in scWAT and protein concentrations in plasma. scWAT transcript levels (quantified by mRNA sequencing) correlated with corresponding plasma protein concentrations for (a,b) leptin and (c,d) high-molecular-weight adiponectin in both groups at baseline. The results were similar using either Spearman’s or Pearson’s correlations. Black lines and dots, control participants; red lines and dots, men with dysglycaemia. Trend lines with standard error of estimates (SEEs; shaded areas) are presented. RPKM, reads per kilobase per million mapped reads
Plasma adipokines response to exercise
Because SFRP4 mRNA showed the largest change in response to the exercise intervention (i.e. lowest p values) across all applied methods (Table 3), and has a known signalling peptide (Table 3), we investigated the effect of exercise on plasma SFRP4 levels. Whereas scWAT SFRP4 mRNA levels did not correlate to plasma SFRP4 concentrations in control participants (Fig. 9a), a significant correlation was seen in men with dysglycaemia (Fig. 9b). Furthermore, no change in plasma SFRP4 concentration was seen in the control group after 12 weeks of exercise (Fig. 9c), whereas a reduction was observed in men with dysglycaemia.
Analysis of SFRP4. (a) Plasma SFRP4 concentrations did not correlate with SFRP4 levels in scWAT in men with normoglycaemia (control), but did correlate in (b) men with dysglycaemia. The results were similar using either Spearman’s and Pearson’s correlations. Trend lines with standard error of estimates (SEEs; shaded areas) are presented. (c) Plasma SFRP4 concentrations did not change after 12 weeks of exercise in men with normoglycaemia, but decreased in men with dysglycaemia. Black lines and dots, control participants; red lines and dots, men with dysglycaemia. RPKM, reads per kilobase of transcript per million mapped reads. *p<0.05, paired t tests were performed
Skeletal muscle mRNA levels and plasma adipokine levels
No correlations were observed between skeletal muscle mRNA levels and plasma concentrations of leptin, total or high-molecular-weight adiponectin, IL-6 or SFRP4 (data not shown).
Discussion
Our main findings were: (1) a substantial difference between change in scWAT transcript levels in response to 12 weeks of exercise among men with dysglycaemia as compared with the control group; (2) some dysglycaemia-specific transcripts were upregulated, but most transcripts were downregulated after 12 weeks of exercise, and were related to inflammation and macrophages; (3) absolute levels of these immune-related transcripts were elevated in men with dysglycaemia at baseline, but were normalised after 12 weeks of exercise, as compared with control participants; (4) levels of these immune-related transcripts correlated negatively with insulin sensitivity in several comparisons; and (5) exercise responses in the scWAT secretome may be reflected in altered plasma concentration of adipokines such as SFRP4.
Whereas several scientists have described adipose tissue responses to different weight-loss regimens in a variety of human populations and animal models [15, 24, 35, 52], there is a lack of studies on the response to exercise, and especially in reporting results after a long-term period of exercise in humans [15, 20, 21]. One study applied microarrays on scWAT before and after 6 months of an exercise intervention in 14 postmenopausal women and found no apparent alterations in transcript levels [24]. Another study among 25 obese, premenopausal women revealed no changes in the mRNA levels of LEP, ADIPOQ, IL6 and TNF after 12 weeks of bicycle ergometer training [22]. Although large differences exist between these studies and ours with respect to exercise protocols, the sex, age, BMI and ethnicity of the participants and the applied technologies, the most important difference may be that the women in these earlier studies were normoglycaemic and insulin sensitive. Hence, the results were in line with those seen among our normoglycaemic and insulin-sensitive control group.
One striking observation in our data is the effect of long-term exercise on reducing levels of inflammatory mRNAs in scWAT among the overweight, dysglycaemic participants. Our pathway analysis and assessment of high-specificity transcript markers of scWAT cell populations [45] indicated less immune cell infiltration, especially of leucocytes and macrophages, after 12 weeks of the exercise intervention in men with dysglycaemia. This is in line with previous studies suggesting that exercise may reduce immune cell infiltration in the stromal vascular fraction of adipose tissue and positively influence adipose tissue macrophage phenotypes [17, 18, 23]. However, these studies focused on visceral adipose tissue [17, 18, 23], not scWAT.
It should be noted that the different transcriptional alterations in scWAT between the two groups in our study occurred without significant changes in diet, BMI, plasma fasting insulin or C-peptide (Table 1). There were also similar reductions in the amount of scWAT and visceral adipose tissue [26] and similar increases in GIR and \( \dot{V}{\mathrm{O}}_{2\mathrm{max}} \) between the two groups (Table 1). Thus, we can only speculate as to why exercise responses in scWAT differed between the two groups, but we suggest that it might relate to differences in fat tissue at baseline (Table 1). Our men with dysglycaemia had more scWAT (Table 1) and increased scWAT inflammation-related transcript levels (Figs. 3 and 6) compared with the control group at baseline. Whereas reduced amounts of scWAT after 12 weeks of exercise in men with dysglycaemia might counteract inflammation-responses associated with expanded adipose tissue [5, 6], no such alterations occurred in normal-weight men, perhaps because their scWAT only changed within a physiological range. We analysed scWAT transcript responses to weight loss (mostly fat mass) and hyperinsulinaemia (insulin infusion for 3 h) in obese and insulin-resistant humans [47, 48]. These analyses demonstrated that some transcripts responding to 12 weeks of exercise intervention in men with dysglycaemia also responded to fat loss or hyperinsulinaemia. Thus, for these transcripts, the observed responses in our data might not be due to exercise per se, but rather due to loss of fat mass and/or improved insulin sensitivity. However, for the majority of transcripts (Table 3), there may be a direct effect of long-term exercise and alleviation of insulin resistance in men with dysglycaemia.
Although human data are generally lacking regarding scWAT and exercise, reports from experiments in rodents have demonstrated profound effects of exercise on scWAT [4]. Transplanting scWAT from mice performing 11 days of cage wheel exercise into sedentary mice dramatically improved glucose tolerance [4]. This effect lasted for 9 days and was related to increased skeletal muscle and intrascapular brown adipose tissue glucose uptake, perhaps related to alterations in more than 250 putative adipokines, possible mediating the effect on glucose tolerance [4]. Although the relevance of these findings to humans remains uncertain, they are in line with our results showing that especially macrophage-related transcript expression seems to be negatively correlated with insulin sensitivity, and that much of this effect may relate to interplay with adipocytes through, for example, adiponectin (Fig. 6 and ESM Tables 1–6) [53]. Thus, both data from Stanford et al [4] and our study suggest that some of the effects of long-term exercise on insulin sensitivity are mediated by alterations in scWAT.
Previous studies on exercise and adipokines have described minor effects, at least compared with different weight-loss regimes [20]. These observations are in agreement with our study, which revealed modest changes in mRNA levels (Fig. 2) and minor changes in plasma adipokine concentrations (Fig. 9). However, subtle effects might still reflect important adaptations; for example, SFRP4 has been shown to be secreted from human scWAT explants [54], and scWAT is the major contributor to plasma SFRP4 concentrations, correlating with indices of obesity and insulin resistance [55]. A mechanism linking elevated plasma SFRP4 concentration to type 2 diabetes involves inflammation in pancreatic islets and reduced insulin secretion [56].
The main limitation of our study is the small sample size of white men only, providing low statistical power and reduced generalisability. However, we applied state-of-the art technologies on human participants who were overweight and of normal weight, and with and without dysglycaemia, who underwent a strictly controlled high-intensity exercise intervention, and compared several statistical approaches across multiple technological platforms to minimise bias. Moreover, the published literature on adipokines and exercise is limited and incomplete, especially in regard to scWAT.
Conclusion
We discovered a pronounced effect of long-term exercise on scWAT in overweight men with dysglycaemia, whereas small alterations were observed in normal-weight men with normoglycaemia. The effect included normalisation of macrophage-related transcript levels, and was closely related to improved insulin sensitivity. The alterations in scWAT involved several secreted factors, and may be mirrored in alterations of plasma adipokine concentrations, at least for SFRP4. These findings indicate that scWAT may be an important mediator of exercise-induced improvements in insulin sensitivity for individuals at risk of developing type 2 diabetes.
Data availability
The data are available on request from the authors.
Change history
29 November 2022
A Correction to this paper has been published: https://doi.org/10.1007/s00125-022-05841-z
Abbreviations
- scWAT:
-
Subcutaneous white adipose tissue
- GIR:
-
Glucose infusion rate
- MRS:
-
Magnetic resonance spectroscopy
- SFRP4:
-
Secreted frizzled-related protein 4
- \( \dot{V}{\mathrm{O}}_{2\mathrm{max}} \) :
-
Maximal oxygen uptake
References
Knights AJ, Funnell AP, Pearson RC, Crossley M, Bell-Anderson KS (2014) Adipokines and insulin action: A sensitive issue. Adipocyte 3(2):88–96. https://doi.org/10.4161/adip.27552
Drevon CA (2005) Fatty acids and expression of adipokines. Biochim Biophys Acta 1740(2):287–292. https://doi.org/10.1016/j.bbadis.2004.11.019
Görgens SW, Eckardt K, Jensen J, Drevon CA, Eckel J (2015) Exercise and regulation of adipokine and myokine production. In: Claude B (ed) Progress in molecular biology and translational science. Academic Press, Cambridge, pp 313–336
Stanford KI, Middelbeek RJ, Goodyear LJ (2015) Exercise effects on white adipose tissue: beiging and metabolic adaptations. diabetes 64(7):2361–2368. https://doi.org/10.2337/db15-0227
Boutens L, Stienstra R (2016) Adipose tissue macrophages: going off track during obesity. Diabetologia 59(5):879–894. https://doi.org/10.1007/s00125-016-3904-9
McLaughlin T, Deng A, Yee G et al (2010) Inflammation in subcutaneous adipose tissue: relationship to adipose cell size. Diabetologia 53(2):369–377. https://doi.org/10.1007/s00125-009-1496-3
Bergmann K, Sypniewska G (2013) Diabetes as a complication of adipose tissue dysfunction. Is there a role for potential new biomarkers? Clin Chem Lab Med 51:177–185
Dolinkova M, Dostalova I, Lacinova Z et al (2008) The endocrine profile of subcutaneous and visceral adipose tissue of obese patients. Mol Cell Endocrinol 291(1-2):63–70. https://doi.org/10.1016/j.mce.2008.05.001
Huber J, Kiefer FW, Zeyda M et al (2008) CC chemokine and CC chemokine receptor profiles in visceral and subcutaneous adipose tissue are altered in human obesity. J Clin Endocrinol Metab 93(8):3215–3221. https://doi.org/10.1210/jc.2007-2630
Samaras K, Botelho NK, Chisholm DJ, Lord RV (2010) Subcutaneous and visceral adipose tissue gene expression of serum adipokines that predict type 2 diabetes. Obesity (Silver Spring) 18(5):884–889. https://doi.org/10.1038/oby.2009.443
Anderssen S, Holme I, Urdal P, Hjermann I (1995) Diet and exercise intervention have favourable effects on blood pressure in mild hypertensives: the Oslo Diet and Exercise Study (ODES). Blood Press 4(6):343–349. https://doi.org/10.3109/08037059509077619
Anderssen SA, Hjermann I, Urdal P, Torjesen PA, Holme I (1996) Improved carbohydrate metabolism after physical training and dietary intervention in individuals with the “atherothrombogenic syndrome”. Oslo Diet and Exercise Study (ODES). A randomized trial. J Intern Med 240(4):203–209. https://doi.org/10.1046/j.1365-2796.1996.22848000.x
Torjesen PA, Birkeland KI, Anderssen SA, Hjermann I, Holme I, Urdal P (1997) Lifestyle changes may reverse development of the insulin resistance syndrome. The Oslo Diet and Exercise Study: a randomized trial. Diabetes Care 20(1):26–31. https://doi.org/10.2337/diacare.20.1.26
Anderssen SA, Holme I, Urdal P, Hjermann I (1998) Associations between central obesity and indexes of hemostatic, carbohydrate and lipid metabolism. Results of a 1-year intervention from the Oslo Diet and Exercise Study. Scand J Med Sci Sports 8(2):109–115
Thompson D, Karpe F, Lafontan M, Frayn K (2012) Physical activity and exercise in the regulation of human adipose tissue physiology. Physiol Rev 92(1):157–191. https://doi.org/10.1152/physrev.00012.2011
Stanford KI, Lynes MD, Takahashi H et al (2018) 12,13-diHOME: an exercise-induced lipokine that increases skeletal muscle fatty acid uptake. Cell Metab 27(5):1111–1120.e1113. https://doi.org/10.1016/j.cmet.2018.03.020
Karsten K, Frank CM, Klaus E, Robert R (2014) Immune and inflammatory signaling pathways in exercise and obesity. Am J Lifestyle Med 10:268–279
Goh J, Goh KP, Abbasi A (2016) Exercise and adipose tissue macrophages: new frontiers in obesity research? Front Endocrinol 7:65
Sakurai T, Ogasawara J, Shirato K et al (2017) Exercise training attenuates the dysregulated expression of adipokines and oxidative stress in white adipose tissue. Oxidative Med Cell Longev 2017:9410954
Campbell KL, Landells CE, Fan J, Brenner DR (2017) A systematic review of the effect of lifestyle interventions on adipose tissue gene expression: implications for carcinogenesis. Obesity (Silver Spring) 25(Suppl 2):S40–S51. https://doi.org/10.1002/oby.22010
Van Pelt DW, Guth LM, Horowitz JF (2017) Aerobic exercise elevates markers of angiogenesis and macrophage IL-6 gene expression in the subcutaneous adipose tissue of overweight-to-obese adults. J Appl Physiol 123(5):1150–1159. https://doi.org/10.1152/japplphysiol.00614.2017
Polak J, Klimcakova E, Moro C et al (2006) Effect of aerobic training on plasma levels and subcutaneous abdominal adipose tissue gene expression of adiponectin, leptin, interleukin 6, and tumor necrosis factor alpha in obese women. Metab Clin Exp 55(10):1375–1381. https://doi.org/10.1016/j.metabol.2006.06.008
Catenacci VA, Wyatt HR (2007) The role of physical activity in producing and maintaining weight loss. Nat Clin Pract Endocrinol Metab 3(7):518–529. https://doi.org/10.1038/ncpendmet0554
Campbell KL, Foster-Schubert KE, Makar KW et al (2013) Gene expression changes in adipose tissue with diet- and/or exercise-induced weight loss. Cancer Prev Res 6(3):217–231. https://doi.org/10.1158/1940-6207.CAPR-12-0212
Pourteymour S, Eckardt K, Holen T et al (2017) Global mRNA sequencing of human skeletal muscle: search for novel exercise-regulated myokines. Molecular metabolism 6(4):352–365. https://doi.org/10.1016/j.molmet.2017.01.007
Langleite TM, Jensen J, Norheim F et al (2016) Insulin sensitivity, body composition and adipose depots following 12 w combined endurance and strength training in dysglycemic and normoglycemic sedentary men. Arch Physiol Biochem 122(4):167–179. https://doi.org/10.1080/13813455.2016.1202985
Andersen LF, Nes M, Lillegaard IT, Sandstad B, Bjorneboe GE, Drevon CA (1995) Evaluation of a quantitative food frequency questionnaire used in a group of Norwegian adolescents. Eur J Clin Nutr 49(8):543–554
Andersen LF, Solvoll K, Johansson LR, Salminen I, Aro A, Drevon CA (1999) Evaluation of a food frequency questionnaire with weighed records, fatty acids, and alpha-tocopherol in adipose tissue and serum. Am J Epidemiol 150(1):75–87. https://doi.org/10.1093/oxfordjournals.aje.a009921
DeFronzo RA, Tobin JD, Andres R (1979) Glucose clamp technique: a method for quantifying insulin secretion and resistance. Am J Phys 237:E214–E223
Stefan D, Cesare FD, Andrasescu A et al (2009) Quantitation of magnetic resonance spectroscopy signals: the jMRUI software package. Meas Sci Technol 20(10):104035. https://doi.org/10.1088/0957-0233/20/10/104035
Carvalho BS, Irizarry RA (2010) A framework for oligonucleotide microarray preprocessing. Bioinformatics 26(19):2363–2367. https://doi.org/10.1093/bioinformatics/btq431
Trapnell C, Roberts A, Goff L et al (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7(3):562–578. https://doi.org/10.1038/nprot.2012.016
Li H, Handsaker B, Wysoker A et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4):357–359. https://doi.org/10.1038/nmeth.1923
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842. https://doi.org/10.1093/bioinformatics/btq033
Pyl PT, Anders S, Huber W (2014) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140. https://doi.org/10.1093/bioinformatics/btp616
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11(10):R106. https://doi.org/10.1186/gb-2010-11-10-r106
Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP (2007) GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 23(23):3251–3253. https://doi.org/10.1093/bioinformatics/btm369
Benjamini Y (2010) Discovering the false discovery rate. J R Stat Soc Ser B Stat Methodol 72(4):405–416. https://doi.org/10.1111/j.1467-9868.2010.00746.x
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8(10):785–786. https://doi.org/10.1038/nmeth.1701
Käll L, Krogh A, Sonnhammer ELL (2007) Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res 35:W429–W432. https://doi.org/10.1093/nar/gkm256
Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300(4):1005–1016. https://doi.org/10.1006/jmbi.2000.3903
Horton P, Park K-J, Obayashi T et al (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35:W585–W587. https://doi.org/10.1093/nar/gkm259
Ehrlund A, Acosta JR, Bjork C et al (2017) The cell-type specific transcriptome in human adipose tissue and influence of obesity on adipocyte progenitors. Scientific data 4:170164. https://doi.org/10.1038/sdata.2017.164
Hill AA, Reid Bolus W, Hasty AH (2014) A decade of progress in adipose tissue macrophage biology. Immunol Rev 262(1):134–152. https://doi.org/10.1111/imr.12216
Magkos F, Fraterrigo G, Yoshino J et al (2016) Effects of moderate and subsequent progressive weight loss on metabolic function and adipose tissue biology in humans with obesity. Cell Metab 23(4):591–601. https://doi.org/10.1016/j.cmet.2016.02.005
Soronen J, Laurila PP, Naukkarinen J et al (2012) Adipose tissue gene expression analysis reveals changes in inflammatory, mitochondrial respiratory and lipid metabolic pathways in obese insulin-resistant subjects. BMC Med Genet 5(1):9. https://doi.org/10.1186/1755-8794-5-9
Ritchie ME, Smyth GK, Phipson B et al (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47–e47. https://doi.org/10.1093/nar/gkv007
Team RC (2018) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Hillier TA, Rousseau A, Lange C et al (2006) Practical way to assess metabolic syndrome using a continuous score obtained from principal components analysis. Diabetologia 49(7):1528–1535. https://doi.org/10.1007/s00125-006-0266-8
Mathur SK, Jain P, Mathur P (2011) Microarray evidences the role of pathologic adipose tissue in insulin resistance and their clinical implications. J Obes 2011:16
Luo Y, Liu M (2016) Adiponectin: a versatile player of innate immunity. J Mol Cell Biol 8(2):120–128. https://doi.org/10.1093/jmcb/mjw012
Ehrlund A, Mejhert N, Lorente-Cebrian S et al (2013) Characterization of the Wnt inhibitors secreted frizzled-related proteins (SFRPs) in human adipose tissue. J Clin Endocrinol Metab 98(3):E503–E508. https://doi.org/10.1210/jc.2012-3416
Garufi G, Seyhan AA, Pasarica M (2015) Elevated secreted frizzled-related protein 4 in obesity: a potential role in adipose tissue dysfunction. Obesity (Silver Spring) 23(1):24–27. https://doi.org/10.1002/oby.20915
Mahdi T, Hanzelmann S, Salehi A et al (2012) Secreted frizzled-related protein 4 reduces insulin secretion and is overexpressed in type 2 diabetes. Cell Metab 16(5):625–633. https://doi.org/10.1016/j.cmet.2012.10.009
Acknowledgements
We thank Å. Halsne, G. Vinje, K. E. Jahnsen, A. Heck, B. Nellemann (Department of Endocrinology, Oslo University Hospital), A. R. Enget (Department of Nutrition, University of Oslo), T. I. Gloppen, T. Dalen, H. Moen, M. A. Dahl, G. Grøthe, K. A Krog, Ø. Skattebo, E. Johansen, D. S. Tangen, K. K. Jensen, H. K. Stadheim, E. N. Rise (Norwegian School of Sport Sciences) and the Norwegian Sequencing Centre.
Funding
This work was supported by grants from the Institute of Basic Medical Sciences, UiO, the Johan Throne-Holst Foundation for Nutrition Research, the Freia Medical Research Foundation, the ‘Functional Genomics’ and ‘Infrastructure’ programmes of the Research Council of Norway, the EU-financed FP7 project (NutriTech grant agreement no.: 289511) and the South-Eastern Regional Health Authorities.
Author information
Authors and Affiliations
Contributions
SL analysed and prepared the data, and wrote the first draft of the manuscript. All authors interpreted the data and reviewed, revised and approved the manuscript. KIB and CAD initiated, designed and supervised the study. CAD is the guarantor of this work.
Corresponding author
Ethics declarations
The authors declare that there is no duality of interest associated with this manuscript.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: The authors regret that they failed to obtain permission to acknowledge J. Jensen (Norwegian School of Sport Sciences).
Electronic supplementary material
ESM
(PDF 394 kb)
Rights and permissions
About this article
Cite this article
Lee, S., Norheim, F., Langleite, T.M. et al. Effects of long-term exercise on plasma adipokine levels and inflammation-related gene expression in subcutaneous adipose tissue in sedentary dysglycaemic, overweight men and sedentary normoglycaemic men of healthy weight. Diabetologia 62, 1048–1064 (2019). https://doi.org/10.1007/s00125-019-4866-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-019-4866-5