Abstract
Aims/hypothesis
To study the heritability and familiality of type 2 diabetes and related quantitative traits in families from the Botnia Study in Finland.
Methods
Heritability estimates for type 2 diabetes adjusted for sex, age and BMI are provided for different age groups of type 2 diabetes and for 34 clinical and metabolic traits in 5,810 individuals from 942 families using a variance component model (SOLAR). In addition, family means of these traits and their distribution across families are calculated.
Results
The strongest heritability for type 2 diabetes was seen in patients with age at onset 35–60 years (h 2 = 0.69). However, including patients with onset up to 75 years dropped the h 2 estimates to 0.31. Among quantitative traits, the highest h 2 estimates in all individuals and in non-diabetic individuals were seen for lean body mass (h 2 = 0.53–0.65), HDL-cholesterol (0.52–0.61) and suppression of NEFA during OGTT (0.63–0.76) followed by measures of insulin secretion (insulinogenic index [IG30] = 0.41–0.50) and insulin action (insulin sensitivity index [ISI] = 0.37–0.40). In contrast, physical activity showed rather low heritability (0.16–0.18), whereas smoking showed strong heritability (0.57–0.59). Family means of these traits differed two- to fivefold between families belonging to the lowest and highest quartile of the trait (p < 0.00001).
Conclusions/interpretation
To detect stronger genetic effects in type 2 diabetes, it seems reasonable to restrict inclusion of patients to those with age at onset 35–60 years. Sequencing of families with extreme quantitative traits could be an important next step in the dissection of the genetics of type 2 diabetes.
Similar content being viewed by others
Introduction
Type 2 diabetes is a multifactorial polygenic disease, which most likely results from the interaction between several genetic and environmental factors. The genetic nature of the disease is supported by stronger heritability in monozygotic than in dizygotic twins [1–3], a sibling relative risk (λ S) of about 3 [4] and high prevalence in certain ethnic groups [5]. A family history of diabetes together with a BMI ≥ 30 kg/m2 and fasting plasma glucose ≥5.5 mmol/l is associated with a 16-fold increased risk of future type 2 diabetes [4]. In support of this, non-diabetic first-degree relatives of patients with type 2 diabetes show early defects in insulin secretion and action [6]. On the other hand, family members also share an environment, and environmental factors such as excess food intake and poor physical activity are important risk factors for type 2 diabetes. Notably, family members of patients with type 2 diabetes are also characterised by poor physical fitness [6]. However, little, if any, information exists on heritability of type 2 diabetes in non-twin families. As environmental factors might contribute more to phenocopies, i.e. non-genetically determined phenotype, of the disease in older age groups, it would be important to define the age group with the highest heritability of the disease.
Recent genome-wide association studies (GWAS) have identified approximately 50 loci associated with type 2 diabetes and glucose and/or insulin levels [7–9], but these variants can explain <15% of the λS value. It has been proposed that intermediate and rare variants with stronger effects might explain the missing heritability of type 2 diabetes [10] but their identification in classical case–control studies of outbred populations might be difficult. One appealing alternative is to apply new generation sequencing to families showing high heritability of an intermediate quantitative trait.
To provide this information we estimated the heritability and familiality of type 2 diabetes and 34 quantitative traits in 5,810 individuals from 942 families participating in the Botnia Study in Finland. The data demonstrate the importance of restricting the search for genetic variants contributing to type 2 diabetes to middle-aged people between 35 and 60 years.
Methods
Subjects
The Botnia Study was initiated in 1990 on the west coast of Finland with the aim of identifying early metabolic defects characteristic of, and genetic factors contributing to, type 2 diabetes [11]. All patients with type 2 diabetes and their family members from five healthcare centres (Närpes, Malax-Korsnäs, Korsholm, Jakobstad and Vasa) were invited to participate in the study.
Family information was obtained on 25,962 individuals from 1,131 families. For the heritability estimates of quantitative traits and type 2 diabetes, we restricted the analysis to families with phenotype information from at least two members, which resulted in 5,810 individuals from 942 families with an average family size of 6.16. Of these, 1,707 had type 2 diabetes. Diabetes was diagnosed using WHO 1998 criteria. Patients with GAD antibodies or known MODY mutations were excluded.
Clinical and metabolic characteristics are presented for all individuals and non-diabetic individuals 16 years or older, as well as all individuals and non-diabetic individuals between 16 and 65 years of age (Table 1).
Written informed consent was obtained from all study participants. The study protocols were approved by local ethics committees, as well as by the ethics committees of Helsinki University Central Hospital and Lund University.
Measurements
After an overnight fast, a polyethylene catheter was placed in an antecubital vein and blood samples were drawn for the analysis of fasting plasma glucose, serum insulin, cholesterol, triacylglycerol, HDL-cholesterol, apolipoproteins A1, A2 and B and NEFA concentrations. Height (to the nearest cm without shoes) and weight (to the nearest kg in light indoor clothing) were recorded. BMI was calculated as weight/height2 (kilograms per square metre). Waist circumference (to the nearest cm without clothes) was measured with a non-elastic soft tape on standing participants midway between the lowest rib and the iliac crest. Hip circumference (to the nearest cm without clothes) was measured over the widest part of the gluteal region. As a measure of abdominal obesity, WHR was calculated. Lean body mass was estimated by a device (Futrex, Gaithersburg, MD, USA) measuring absorption of infrared light in subcutaneous tissue. Blood pressure was measured as the mean of two recordings with the patient in the supine position [11].
For the OGTT, participants ingested 75 g dextrose and blood samples for the measurements of plasma glucose and serum insulin concentrations were drawn at −5, 0, 30, 60 and 120 min.
Plasma glucose was measured with a glucose oxidase method (Beckman Glucose Analyzer, Beckman Instruments, Fullerton, CA, USA) and serum insulin by an enzyme immunoassay (DAKO, Cambridgeshire, UK) with an interassay CV of 7.5. Serum total cholesterol, HDL-cholesterol and triacylglycerol concentrations were measured on a Cobas Mira analyser (Hoffman–LaRoche, Basel, Switzerland) and LDL-cholesterol concentration was calculated using the Friedewald formula. The serum concentrations of apolipoproteins A1, A2 and B were measured by immunochemical assays (Orion Diagnostica, Espoo, Finland), and that of NEFA by an enzymatic colorimetric method (Wako Chemicals, Neuss, Germany).
Insulin resistance was estimated as the HOMA of insulin resistance (HOMA-IR) [12], and insulin sensitivity from the OGTT was calculated as the Matsuda insulin sensitivity index (ISI) [13]. Beta cell function was assessed as the ratio of incremental insulin (ΔI 30) to glucose (ΔG 30) responses during the first 30 min of the OGTT (ΔI/ΔG = [I 30 − I 0]/[G 30 − G 0]), also called the insulinogenic index (IG30), or as the HOMA beta cell index [12]. The disposition index (DI) was used to adjust insulin secretion for the degree of insulin resistance (IG30 × HOMA-IR). Urinary albumin concentration was measured by radioimmunoassay (Pharmacia-Upjohn, Uppsala, Sweden). Urine for the measurement of AER was collected either during the OGTT or overnight. Microalbuminuria was defined as AER >20 μg/min.
At the baseline examination, a structured questionnaire was used to collect the data on smoking habits and self-reports of physical activity during work and on the way to work [14]. The physical activity data were converted into metabolic units (MET), which estimate the relative intensity of the activity as the energy needed during exercise compared with the resting energy expenditure (1 MET approximately equals the energy expenditure of 4.18 kJ [1 kcal]/kg of body weight per h or oxygen uptake of 3.5 ml kg−1 min−1). Physical activity at work was estimated with a seven point scale ranging from no activity and no work (1.5 MET) to very heavy manual work (10.0 MET). Exercise activity on the way to work was graded into three levels: travel by motor vehicle (1.5 MET), on foot (3.5 MET) and by bicycle (5 MET). The sum of the different MET values was used to grade the total physical activity of the participants [4].
Statistics
Unless otherwise stated, values are presented as mean ± SD. Non-symmetrically distributed variables were log-transformed before analysis. Heritability (h 2) estimates the extent to which genetic factors contribute to the quantitative traits. The SOLAR software package provides a maximum likelihood estimate of heritability taking into account additive genetic effects and residual errors [15]. All quantitative trait analyses were adjusted for age, sex, the interaction between age and sex, age2 and affection status (type 2 diabetes). In addition, all traits except BMI were adjusted for BMI. The covariates not found to be significant after covariate screening were removed from the final models. Heritability thus represents the proportion of phenotypic variance due to additive genetic effects after adjustment for covariates.
Heritability of type 2 diabetes was estimated using a liability threshold model implemented in SOLAR [16]. In addition to the common covariates (age, sex, age × sex, age2 and BMI), we added a SNP in TCF7L2, rs7903146, as a covariate for the analysis of type 2 diabetes.
Sibling relative risk (λ S) was calculated as risk of diabetes in siblings of patients with type 2 diabetes compared with the background population following the method proposed by Olson and Cordell [17].
Differences in Pearson correlation coefficients between parent–son and parent–daughter pairs were evaluated using the corcor function (Goldstein, Qualitas, Brighton, MA, USA) for testing dependent correlation coefficients in STATA (Stata, TX, USA).
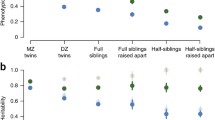
For calculation of family means of the quantitative traits, we included only families with measurements available in at least three individuals. Data are shown as histograms and ranked mean (min–max) values from the lowest to the highest across the families (see Electronic supplementary material [ESM] Figs 1 and 2). Ranked mean (range) values for the 20 families with lowest and highest values for height, BMI, IG30, ISI, HDL-cholesterol and 2 h NEFA during OGTT are shown in Fig. 1.
Mean values of height (a), BMI (b), insulinogenic index (c), insulin sensitivity index (d), HDL-cholesterol (e) and NEFA concentrations (f) at 120 min of OGTT for the 20 families from the lower and 20 families from the upper extreme tails of the distributions. The vertical lines represent minimum and maximum values within the families
Results
The clinical and metabolic characteristics of the 5,810 individuals aged 16 years or older from 942 families are shown in Table 1; of these, 1,707 had type 2 diabetes. Sex distribution was similar among all and non-diabetic individuals (both 46% males). Data are separately shown for individuals below the age of 65 years. As expected, diabetes (present in about 30% of all individuals) had the strongest influence on glucose, HbA1c and triacylglycerol concentrations as well as on AER.
Heritability of type 2 diabetes
Heritability estimates were adjusted for age, sex, their interaction, age2 and BMI (Table 2). Heritability was rather low when individuals in all age groups were considered (h 2 = 0.25). The highest heritability for type 2 diabetes was seen in the age group 35–60 years (0.69; p < 0.0001) (Table 2, ESM Table 1); this dropped markedly to 0.31 when the upper age was increased to 75 years. Clinical characteristics and heritability estimates for quantitative traits in this age group are given in ESM Tables 2 and 3. The sibling relative risk, i.e. the risk of diabetes in siblings of patients with type 2 diabetes compared with the risk in the background population (λ S), was clearly higher than previously estimated from twin studies, i.e. 7.95 using all individuals 16 years or older. We also tested whether the SNP rs7903146 in the gene showing the strongest association with type 2 diabetes, TCF7L2, influenced the heritability estimates. Although h 2 dropped from 0.72 to 0.64 after adjustment for the TCF7L2 SNP in the age group 35–60 years, this difference did not reach statistical significance given the large variance in the estimates.
Heritability of anthropometric measurements, blood pressure and AER
The highest heritability was seen for height (Table 3) (ranging from 0.81 to 0.88), followed by lean body mass (0.53–0.69), BMI (0.46–0.53), waist circumference (0.42–0.50) and waist/height2 (0.47–0.49), whereas h 2 for WHR was rather low ranging from 0.27 to 0.32. As reported previously [18], there was a modest heritability for AER in all as well as in non-diabetic individuals (0.35 and 0.30 respectively).
Heritability of glycaemia, insulin secretion and insulin sensitivity
As predicted, heritability estimates for glycaemia performed poorly in all individuals due to inclusion of diabetic individuals (Table 3). Fasting glucose concentrations showed higher heritability than 2 h glucose concentrations in the non-diabetic individuals (0.58 vs 0.32), whereas the opposite was seen for fasting and 2 h insulin concentrations (0.41 vs 0.34). Heritability estimates for beta cell function (IG30, HOMA beta cell index) and insulin sensitivity/resistance (ISI, HOMA-IR) were in the same range, suggesting that 40–50% of the variance in these measurements can be explained by genetic factors in non-diabetic individuals.
Heritability of lipids and lipoproteins
The diabetic state had little influence on heritability of cholesterol concentrations but somewhat more on HDL-cholesterol, triacylglycerol and apolipoprotein concentrations (Table 3). Of all lipids, HDL-cholesterol concentrations showed the highest h 2 (0.52–0.62). Intriguingly, the highest heritability in all and in non-diabetic individuals was seen for suppression of NEFA concentrations at 2 h of the OGTT (0.63–0.76).
Heritability of physical activity and smoking
We also estimated heritability for two traits considered as environmental, physical activity and smoking. The heritability estimates for physical activity were low (0.16–0.21), supporting the view that physical activity is mostly influenced by environmental factors. In contrast, heritability estimates for smoking were very high (0.57–0.63) emphasising a large contribution of genetic factors to smoking habits (Table 3).
Parent–offspring correlations in quantitative traits
To investigate differences in transmission between sexes, we calculated correlations between parents and offspring for 31 different traits. For five traits we observed a difference in parent–offspring correlation between fathers and sons and mothers and sons, and for nine between father–daughter and mother–daughter pairs (ESM Table 4). Cholesterol differed in all sex-specific parent-of-origin correlations.
Distribution of quantitative traits across families
Distribution of family means for height, BMI, IG30, ISI, HDL-cholesterol and NEFA concentrations at 2 h of the OGTT are shown as histograms in ESM Fig. 1. The corresponding family means and ranges for 20 families from the lower (5%) and 20 families from the upper tails of the trait distribution are shown in Fig. 1 and for all families in ESM Fig. 2. The family means for other traits from the families from the lower and upper tails of distribution are shown in ESM Fig. 3.
Although most traits followed a symmetrical distribution with an equal proportion of extremes at both ends of distribution (ESM Fig. 1), suppression of NEFA during OGTT showed a skewed distribution with a large number of families with very low NEFA values, suggesting high sensitivity to suppression of NEFA during OGTT.
Although there was a large intra-familial variance in height, BMI, IG30, ISI, HDL-cholesterol and NEFA 2 h, the interfamily differences were larger. For HDL-cholesterol the difference between the families with lowest and highest mean HDL-cholesterol was twofold. This is also illustrated by segregation of HDL-cholesterol in two families from each extreme of the distribution (ESM Fig. 4).
Discussion
To our knowledge, this is the first time heritability estimates for type 2 diabetes as a dichotomous trait have been presented in addition to estimates of heritability for anthropometric, metabolic and environmental traits in families rather than in twins. The large family-based Botnia Study is well suited for this purpose. The heritability estimates for 34 clinical, metabolic and environmental traits in 5,810 individuals from 942 families showed that (a) the highest heritability for type 2 diabetes is restricted to middle-aged people aged 35–60 years and decreases markedly if the upper age limit is increased to 75 years, (b) the sibling risk for type 2 diabetes (λ S) was much higher than estimated from twin studies (8 vs 3), (c) the strongest heritabilities for quantitative traits were seen for height, lean body mass, fasting HDL-cholesterol concentrations and NEFA concentrations at 2 h of an OGTT and (d) distribution of these traits across families should allow selection of families with extreme trait values for next-generation sequencing studies.
The observed large differences in heritability estimates of type 2 diabetes between different age groups send a clear message that genetic studies on type 2 diabetes might benefit from restricting inclusion of individuals to the age group 35–60 years, with the upper age limit possibly being extended to 65. Unfortunately, this has not been the case in most GWAS studies on type 2 diabetes [7, 19, 20]. The proportion of the trait variance not attributed to genetic factors increased when including individuals with late onset diabetes. Although a recent meta-analysis by Voight et al. [9] did not demonstrate any heterogeneity based on age at onset, it might be difficult to identify such an effect in cohorts ascertained using different criteria. Also, the age window may differ between different ethnic groups as onset of diabetes seems to be earlier in patients from the Middle East than in patients from Scandinavia [21]. Inclusion of the strongest type 2 diabetes-associated SNP, rs7903146 in the TCF7L2 gene, in the analysis did not result in a significant decrease in the heritability estimates in the age group 35–60 years. Whether this reflects insufficient power due to large variance in the estimates or a true negative finding remains to be shown. Another intriguing finding was that the sibling risk of diabetes (λ S) was much higher than obtained in twin studies (8 vs 3). This may not be that surprising as the current data were obtained from families with an average family size of 6.16 and most twin studies did not take into account family history of diabetes. The high λ S in families is in keeping with data from a Swedish study using the multigeneration registry [22]. In this study the sibling-relative risk of developing diabetes was as high as 39 if the person had two siblings with diabetes. These findings can be interpreted in two ways. One would be that if we have underestimated heritability estimates of type 2 diabetes, current GWAS studies have been able to identify even less of the heritability than the present view of 10–15% assumes. The other would be that we have been searching for genes in the wrong place, i.e. in outbred populations with little evidence for heritability of type 2 diabetes.
The current heritability estimate for type 2 diabetes obtained in a large number of families using a variance component analysis model SOLAR provides an alternative to the traditional λ S estimate of heritability, and may be better suited to testing the proportion of heritability accounted for by known type 2 diabetes-associated genetic variants as it is obtained in the same families where the variants are genotyped.
The heritabilities of anthropometric traits such as height and BMI were in the same range as reported in other studies [23–28]. However, the observation that lean body mass is strongly heritable is new and supports previous findings that bone mass is genetically determined in twin studies [29, 30]. It further suggests that lean body mass (muscle and bone) may be under stronger genetic control than fat mass.
In contrast to findings from twin studies [28, 31] and one family study [24] the OGTT-based measures of insulin secretion (IG30) and insulin sensitivity (ISI) showed a similar degree of heritability (40–50%), which was higher than seen for the corresponding HOMA measurements. Family means of these traits indicated clear clustering of insulin secretion and action in families with greater intra-family variability for measures of insulin sensitivity than for measures of insulin secretion (Fig. 1). The families at both ends of the distribution should be attractive targets for next-generation sequencing efforts. We could confirm previous studies showing high heritability for HDL-cholesterol concentrations in families [23, 26, 27] and add new information on heritability of apolipoproteins in families. In line with a previous study in twins [32], both the HDL-associated apolipoproteins A1 and A2 showed a high degree of heritability in these families. Most interestingly, the highest heritability of any metabolic trait was seen for NEFA during the OGTT. Postprandial NEFA concentrations most likely reflect turnover, lipolysis and fractional esterification of the NEFA pool (NEFA turnover adjusted for the fat mass). This finding also goes along with a report showing impaired suppression of postprandial triacylglycerol concentrations in first-degree relatives of patients with type 2 diabetes [33]. Notably, although most family means showed a typical symmetrical distribution, suppression of NEFA during the OGTT was skewed with a number of families being very sensitive to suppression of NEFA during OGTT (ESM Fig. 1).
Finally, heritability of the two apparently behavioural traits of physical activity and smoking provided some intriguing results. Although physical activity showed low heritability (20%), supporting the notion that it mostly represents an environmental trait that can be modified by other environmental factors [34], smoking was a strongly heritable habit (50–60%) in line with some previous studies [35]. An obvious interpretation of this would be that it should be easier to influence physical activity than smoking by behavioural modifications.
The low heritability for physical activity would at first glance seem at variance with a recent paper from our group showing that physical fitness is lower in first-degree relatives of patients with type 2 diabetes than in individuals without a family history of type 2 diabetes [6]. However, the reduction in physical fitness was seen despite similar amounts of reported physical activity as estimated in the current paper, suggesting that physical fitness is a consequence of a number of factors, physical activity being only one.
We also examined whether heritability estimates would be influenced by sex and transmission from mother or father. For most traits, the correlation was stronger when transmitted from mothers than from fathers. Interestingly, sex-specific parental effects were strongest for cholesterol concentrations. Thereby, these findings using an alternative way of analysing data support our previous findings of sex-specific parent-of-origin effects for many traits [11] and also justify similar analyses for genetic variants influencing these traits [36].
Given that current GWAS have not been able to explain the heritability of type 2 diabetes, future approaches will include means to detect rare and intermediate frequency variants with hopefully stronger effects. As heritability is a measure of segregation of a trait in families it seems reasonable to turn back to families for these efforts rather than to study outbred case–control populations. Exome and whole-genome sequencing of families with extreme type 2 diabetes-related traits [37, 38] provide appealing approaches to find the missing heritability of type 2 diabetes. To provide some background information for such studies, we analysed the distribution of quantitative traits across families. As can be seen from Fig. 1 and ESM Fig. 1, selection of families with extreme values for a trait might provide a fourfold difference in the trait. As many of these traits show 50–70% heritability, we would expect to find an OR of >2 in such sequencing efforts. Another advantage with family studies is that it will be much easier to exclude technical sequencing errors as identified variants can immediately be tested for Mendelian segregation.
In conclusion, to detect stronger genetic effects in type 2 diabetes, it seems reasonable to restrict inclusion of patients to those with age at onset 35–60 years. Sequencing of families with extreme quantitative traits could be an important next step in the dissection of the genetics of type 2 diabetes.
Abbreviations
- DI:
-
Disposition index
- GWAS:
-
Genome-wide association study
- h 2 :
-
Heritability estimate
- HOMA-IR:
-
HOMA of insulin resistance
- IG30 :
-
Insulinogenic index
- ISI:
-
Insulin sensitivity index
- MET:
-
Metabolic unit
References
Kaprio J, Tuomilehto J, Koskenvuo M et al (1992) Concordance for type 1 (insulin-dependent) and type 2 (non-insulin-dependent) diabetes mellitus in a population-based cohort of twins in Finland. Diabetologia 35:1060–1067
Newman B, Selby JV, King MC, Slemenda C, Fabsitz R, Friedman GD (1987) Concordance for type 2 (non-insulin-dependent) diabetes mellitus in male twins. Diabetologia 30:763–768
Poulsen P, Kyvik KO, Vaag A, Beck-Nielsen H (1999) Heritability of type II (non-insulin-dependent) diabetes mellitus and abnormal glucose tolerance—a population-based twin study. Diabetologia 42:139–145
Lyssenko V, Almgren P, Anevski D et al (2005) Predictors of and longitudinal changes in insulin sensitivity and secretion preceding onset of type 2 diabetes. Diabetes 54:166–174
Rimoin DL (1969) Ethnic variability in glucose tolerance and insulin secretion. Arch Intern Med 124:695–700
Isomaa B, Forsen B, Lahti K et al (2010) A family history of diabetes is associated with reduced physical fitness in the Prevalence, Prediction and Prevention of Diabetes (PPP)–Botnia Study. Diabetologia 53:1709–1713
Saxena R, Voight BF, Lyssenko V et al (2007) Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316:1331–1336
Zeggini E, Scott LJ, Saxena R et al (2008) Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 40:638–645
Voight BF, Scott LJ, Steinthorsdottir V et al (2010) Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 42:579–589
Manolio TA, Collins FS, Cox NJ et al (2009) Finding the missing heritability of complex diseases. Nature 461:747–753
Groop L, Forsblom C, Lehtovirta M et al (1996) Metabolic consequences of a family history of NIDDM (the Botnia Study): evidence for sex-specific parental effects. Diabetes 45:1585–1593
Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC (1985) Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28:412–419
Matsuda M, DeFronzo RA (1999) Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care 22:1462–1470
Mälkiä E, Impivaara O, Maatela J, Aromaa A, Heliövaara M, Knekt P (1988) Physical activity of Finnish adults. Publications of the Social Insurance Institution, Turku, ML:80:110
Almasy L, Blangero J (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 62:1198–1211
Duggirala R, Williams JT, Williams-Blangero S, Blangero J (1997) A variance component approach to dichotomous trait linkage analysis using a threshold model. Genet Epidemiol 14:987–992
Olson JM, Cordell HJ (2000) Ascertainment bias in the estimation of sibling genetic risk parameters. Genet Epidemiol 18:217–235
Forsblom CM, Kanninen T, Lehtovirta M, Saloranta C, Groop LC (1999) Heritability of albumin excretion rate in families of patients with type II diabetes. Diabetologia 42:1359–1366
Scott LJ, Mohlke KL, Bonnycastle LL et al (2007) A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316:1341–1345
Zeggini E, Weedon MN, Lindgren CM et al (2007) Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316:1336–1341
Glans F, Elgzyri T, Shaat N, Lindholm E, Apelqvist J, Groop L (2008) Immigrants from the Middle-East have a different form of type 2 diabetes compared with Swedish patients. Diabet Med 25:303–307
Hemminki K, Li X, Sundquist K, Sundquist J (2010) Familial risks for type 2 diabetes in Sweden. Diabetes Care 33:293–297
Austin MA, Edwards KL, McNeely MJ et al (2004) Heritability of multivariate factors of the metabolic syndrome in nondiabetic Japanese Americans. Diabetes 53:1166–1169
Elbein SC, Hasstedt SJ, Wegner K, Kahn SE (1999) Heritability of pancreatic beta-cell function among nondiabetic members of Caucasian familial type 2 diabetic kindreds. J Clin Endocrinol Metab 84:1398–1403
Hsueh WC, Mitchell BD, Aburomia R et al (2000) Diabetes in the Old Order Amish: characterization and heritability analysis of the Amish Family Diabetes Study. Diabetes Care 23:595–601
Lin HF, Boden-Albala B, Juo SH, Park N, Rundek T, Sacco RL (2005) Heritabilities of the metabolic syndrome and its components in the Northern Manhattan Family Study. Diabetologia 48:2006–2012
Mills GW, Avery PJ, McCarthy MI et al (2004) Heritability estimates for beta cell function and features of the insulin resistance syndrome in UK families with an increased susceptibility to type 2 diabetes. Diabetologia 47:732–738
Poulsen P, Levin K, Petersen I, Christensen K, Beck-Nielsen H, Vaag A (2005) Heritability of insulin secretion, peripheral and hepatic insulin action, and intracellular glucose partitioning in young and old Danish twins. Diabetes 54:275–283
Pocock NA, Eisman JA, Hopper JL, Yeates MG, Sambrook PN, Eberl S (1987) Genetic determinants of bone mass in adults. A twin study. J Clin Invest 80:706–710
Smith DM, Nance WE, Kang KW, Christian JC, Johnston CC Jr (1973) Genetic factors in determining bone mass. J Clin Invest 52:2800–2808
Lehtovirta M, Kaprio J, Forsblom C, Eriksson J, Tuomilehto J, Groop L (2000) Insulin sensitivity and insulin secretion in monozygotic and dizygotic twins. Diabetologia 43:285–293
Pietilainen KH, Soderlund S, Rissanen A et al (2009) HDL subspecies in young adult twins: heritability and impact of overweight. Obesity (Silver Spring) 17:1208–1214
Axelsen M, Smith U, Eriksson JW, Taskinen MR, Jansson PA (1999) Postprandial hypertriglyceridemia and insulin resistance in normoglycemic first-degree relatives of patients with type 2 diabetes. Ann Intern Med 131:27–31
Aaltonen S, Ortega-Alonso A, Kujala UM, Kaprio J (2010) A longitudinal study on genetic and environmental influences on leisure time physical activity in the Finnish Twin Cohort. Twin Res Hum Genet 13:475–481
The Tobacco and Genetics Consortium (2010) Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet 42:441–447
Kong A, Steinthorsdottir V, Masson G et al (2009) Parental origin of sequence variants associated with complex diseases. Nature 462:868–874
Lupski JR, Reid JG, Gonzaga-Jauregui C et al (2010) Whole-genome sequencing in a patient with Charcot–Marie–Tooth neuropathy. N Engl J Med 362:1181–1191
Roach JC, Glusman G, Smit AF et al (2010) Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science 328:636–639
Acknowledgements
We thank the Botnia research team in Finland and Sweden for skilful support. The study was supported by grants from the Sigrid Juselius Foundation, the Folkhälsan Research Foundation, Swedish Research Council including a Linné grant, EXODIAB, two EU grants (ENGAGE and CEED3), Novo Nordisk Foundation, Swedish Diabetes Research Foundation, Finnish Diabetes Research Foundation, Foundation for Life and Health in Finland, Helsinki University Central Hospital Research Foundation, Ollqvist Foundation and the Söderberg Foundation. The study has also been supported by the Municipal Heath Care Center and Hospital in Jakobstad, and Health Care Centers in Vasa, Närpes, Malax and Korsholm.
Contribution statement
PA was responsible for the study conception, design and analysis, interpretation of data and drafting the manuscript, and gave final approval of the version of the manuscript to be published. ML, BI, LS, MRT and TT were responsible for phenotyping in the Botnia Study and interpretation of the data, and they revised the manuscript and gave final approval of the version of the manuscript to be published. VL was responsible for the interpretation of data and revising the manuscript, and gave final approval of the version of the manuscript to be published. LG designed and supervised all parts of the study, drafted the report and gave final approval of the version of the manuscript to be published.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Author information
Authors and Affiliations
Consortia
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM Table 1
Heritability of type 2 diabetes in different age groups with covariate sets with and without rs7903146 (PDF 9 kb)
ESM Table 2
Clinical and metabolic characteristics of all individuals 35–60 years and of non-diabetic individuals 35–60 years (PDF 77 kb)
ESM Table 3
Heritability estimates for anthropometric measures, blood pressure, AER, glucose and insulin concentrations, insulin resistance and insulin secretion indexes, HbA1c, lipids, lipoproteins, NEFA, physical activity and smoking in individuals 35–60 years old (PDF 30 kb)
ESM Table 4
Correlation coefficient (r) differences between father–offspring and mother–offspring pairs (PDF 45 kb)
ESM Fig. 1
Distribution of family means of selected traits. Bars represent number of families (PDF 15 kb)
ESM Fig. 2
Mean values of (A) height, (B) BMI, (C) insulinogenic index, (D) insulin sensitivity index, (E) HDL cholesterol and (F) NEFA concentrations at 120 min of OGTT for all families. The vertical lines represent minimum and maximum values within the families (PDF 50 kb)
ESM Fig. 3
Mean values of (A) weight, (B) lean body mass, (C) fat mass, (D) waist circumference, (E) WHR, (F) waist/height2, (G) diastolic blood pressure, (H) systolic blood pressure, (I) albumin excretion rate, (J) fasting P-glucose concentrations, (K) P-glucose concentrations at 120 min of OGTT, (L) HbA1c, (M) fasting S-insulin concentrations, (N) S-insulin concentrations at 120 min of OGTT, (O) disposition index, (P) HOMA-IR, (Q) HOMA beta cell index, (R) total cholesterol, (S) LDL cholesterol, (T) triacylglycerol, (U) triacylglycerol to HDL cholesterol ratio, (V) ApoA1, (W) ApoA2, (X) ApoB and (Y) fasting NEFA concentrations for the 20 families from the lower and 20 families from the upper extreme tails of the distributions. The vertical lines represent minimum and maximum values within the families (PDF 101 kb)
ESM Fig. 4
Two families with segregation of extreme HDL cholesterol values (PDF 8 kb)
Rights and permissions
About this article
Cite this article
Almgren, P., Lehtovirta, M., Isomaa, B. et al. Heritability and familiality of type 2 diabetes and related quantitative traits in the Botnia Study. Diabetologia 54, 2811–2819 (2011). https://doi.org/10.1007/s00125-011-2267-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-011-2267-5