Abstract
Aims/hypothesis
The gene encoding transcription factor 7-like 2 (TCF7L2) has been related to type 2 diabetes in multiple ethnic groups. Here, we investigate whether three single nucleotide polymorphisms (SNPs) in the TCF7L2 gene are associated with an impaired proinsulin:insulin ratio.
Methods
In this study we examined the associations between SNPs rs7901695, rs7903146 and rs12255372 in the TCF7L2 gene and metabolic variables affecting type 2 diabetes in a population-based study of 706 unrelated individuals (47% men and 53% women; aged 35–74 years) from the province of Segovia in central Spain (Castille), including 180 individuals with type 2 diabetes.
Results
The minor allele frequency of rs7901695, rs7903146 and rs12255372 was significantly higher in diabetic patients compared with that in non-diabetic individuals. The T (minor) allele of the variant rs7903146 was significantly associated with a greater OR for type 2 diabetes adjusted for age, sex and BMI in logistic regression analysis: OR 1.29 (95% CI 1.06–1.57, p = 0.01). This risk allele was also associated with an increased proinsulin:insulin ratio after OGTT. Similar results were obtained for the other TCF7L2 SNPs.
Conclusions/interpretation
Our results provide further evidence supporting the belief that the TCF7L2 gene is a major determinant of type 2 diabetes risk in Spain, as in other southern European populations. The association with increased proinsulin:insulin ratio after an OGTT suggests that TCF7L2 might be involved in insulin synthesis and processing.
Similar content being viewed by others
Introduction
Although type 2 diabetes mellitus has a strong genetic basis, until recently most candidate genes for it have shown only modest effects, and the associations have been inconsistent [1].
Recently Grant et al. showed a significant association between a microsatellite marker (DG10S478) located in the gene encoding transcription factor 7-like 2 (TCF7L2) and the risk of type 2 diabetes mellitus [2]. Also TCF7L2 was one of the most significantly associated genes in recent genome-wide association studies [3].
The original association was found in an Icelandic cohort of type 2 diabetes mellitus patients and control individuals and was subsequently replicated in both Danish and American cohorts. The single nucleotide polymorphisms (SNPs) rs7903146 and rs12255372 were in strong linkage disequilibrium, with the DG10S478 microsatellite marker showing similar strong associations. Grant et al. [2] suggested that two SNPs be analysed for replication of their finding in other populations. Since the original publication, numerous published studies have examined this gene, with consistent replication in several other populations [3].
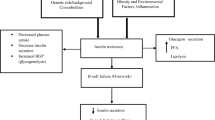
TCF7L2 is widely expressed, including mature pancreatic beta cells as well as peripheral and omental adipocytes. The WNT pathway is important in adipogenesis; thus, a broader physiological role for TCF7L2 in glucose homeostasis is plausible.
TCF7L2 binds to the promoter of the proglucagon gene in gut endocrine L cells, thus potentially acting through glucagon-like peptide 1 to affect insulin secretion. Indeed some, but not all, studies have shown impaired insulin secretion to oral glucose in individuals with the rs7903146 TT genotype [4, 5]. Both Florez et al. [4] and Saxena et al. [5] reported reduced insulin secretion in response to oral glucose in carriers of the risk allele.
Therefore, our study aimed to determine whether three SNPs of the TCF7L2 gene, rs7901695, rs7903146 and rs12255372, are associated with metabolic variables affecting type 2 diabetes, such as those involved in beta cell dysfunction, in a population-based study cohort including 706 unrelated individuals from Spain.
Methods
Population: the Segovia Study, Spain
The Segovia Study was designed as a cross-sectional population-based study of the prevalence of anthropometric and physiological variables related to obesity and other components of the metabolic syndrome. It was conducted in rural and urban areas of the province of Segovia (Community of Castilla-León) in central Spain. In brief, a random sample of 2,992 men and non-pregnant women aged 35–74 years was selected from a target population of 63,417 inhabitants, of whom 1,033 agreed to participate. Individuals with a previous diagnosis of type 1 diabetes were excluded (n = 133) from the sample. All study participants gave written informed consent to participate in the study. The study protocol was approved by the Ethics Committee of the Hospital Clínico San Carlos of Madrid. A random sample (n = 706) was successfully genotyped for the rs7903146 variant of the TCF7L2 gene.
Phenotypic measurements
Anthropometric measurements included BMI (kg/m2) and waist circumference (cm). Obesity was defined as BMI ≥30 kg/m2 (n = 204; 82 men and 122 women). Waist measurements were made with a non-stretchable fibre measuring tape while study participants were standing erect in a relaxed position with both feet together on a flat surface. Waist circumference was measured as the smallest horizontal girth between the costal margins and the iliac crests at minimal respiration.
Systolic and diastolic blood pressures were measured three times on persons in the seated position after 10 min of rest; measurements were read to the nearest even digit by use of a random-zero sphygmomanometer.
A 75 g OGTT was performed in the morning after an overnight fast and interpreted according to the criteria of the American Diabetes Association recommendations. Blood samples were collected from an antecubital vein without compression at 0 and 120 min for glucose, insulin and proinsulin levels.
Plasma glucose was determined in duplicate by a glucose-oxidase method adapted to an autoanalyser (Hitachi 704; Boehringer, Mannheim, Germany). Total cholesterol, triacylglycerol and HDL-cholesterol were determined by enzymatic methods with commercial reagent sets (Boehringer). LDL-cholesterol was calculated by the Friedewald formula.
Serum insulin concentrations were determined by RIA (Human Insulin Specific RIA kit; Linco Research, St Louis, MO, USA) with a lower detection limit of 12 pmol/l; intra- and interassay CV values being <1% and <7.43%, respectively. Cross-reactivity with proinsulin was <2%. Proinsulin serum concentrations were assayed with a sensitive, specific RIA (Human Proinsulin RIA Kit; Linco Research).
The proinsulin:insulin ratio was calculated by dividing the concentration of proinsulin (pmol/l) by insulin (pmol/l). Homeostasis model assessments of insulin resistance (HOMA-IR) and beta cell function (HOMA-B) were calculated as previously described [6].
SNP selection and genotyping
We genotyped three SNPs (rs7901695, rs7903146 and rs12255372) that showed association with type 2 diabetes in the study by Grant et al. [2]. Genotyping of these SNPs was carried out using TaqMan SNP Genotyping Assays (Applied Biosystems, Foster City, CA, USA) and fluorescence was measured using an ABI 7900 Sequence Detection System (Applied Biosystems). To assess genotyping reproducibility, a random ~20% selection of the samples was re-genotyped, with 100% concordance.
Statistical analysis
We compared genotype and allele frequency distributions using the χ 2 test and computed Hardy–Weinberg equilibrium to the expected genotype distribution. Pairwise linkage disequilibrium was estimated using Haploview version 3.2. We used Student’s t test and ANOVA to compare continuous variables (expressed as means [SE]), and the χ 2 test to compare categorical variables. Continuous variables that did not have a Gaussian distribution were log transformed.
Logistic regression analysis was performed to evaluate associations of rs7901695, rs7903146 and rs12255372 SNPs with type 2 diabetes. Adjusted ORs and their 95% CIs were calculated. The null hypothesis was rejected in each statistical test at p < 0.05. Analysis was performed using the Windows SPSS version 15.0 software.
Our study had a power of 76% at p = 0.05 to detect effects with an OR of 1.40 for examined variants.
Results
As expected, the clinical characteristics of participants with and without type 2 diabetes (n = 180) differed for a number of metabolic traits. BMI, waist circumference, systolic and diastolic blood pressure, fasting glucose, insulin, proinsulin, total cholesterol, triacylglycerol and HOMA-IR were higher and HDL-cholesterol concentrations were lower in individuals with type 2 diabetes than in those without (Table 1).
The genotype distribution at all the SNPs did not show any deviation from Hardy–Weinberg equilibrium. The three TCF7L2 SNPs were in high linkage disequilibrium with D΄ = 0.89–0.96 and r 2 = 0.74–0.88; thus, the results of the associations were largely comparable.
Regarding the three SNPs, we observed significant differences in genotypic distributions according to type 2 diabetes. In brief, the minor allele frequency of rs7901695, rs7903146 and rs12255372 showed a significant association with susceptibility to type 2 diabetes. The association with type 2 diabetes was seen for the C allele of the variant rs7901695 (OR 1.59, 95% CI 1.10–2.34, p = 0.02). Similarly, both the T allele of the variant rs12255372 and the T allele of rs7903146 were significantly associated with greater ORs for type 2 diabetes adjusted for age, sex and BMI in logistic regression analysis: OR 1.37 (95% CI 1.05–1.78, p = 0.02) and OR 1.29 (95% CI 1.06–1.57, p = 0.01), respectively.
Because of the results of the associations were similar for the three SNPs, here we describe the results of the most reproducible polymorphism, rs7903146.
In addition to increased type 2 diabetes, the TT genotype was also associated with a higher proinsulin:insulin ratio after an OGTT and a trend towards higher fasting plasma glucose compared with TC and CC genotypes in non-diabetic individuals (Table 2). However, no significant differences were found between these variants in the TCF7L2 gene and obesity measures.
Discussion
We examined the potential associations between type 2 diabetes and SNPs of the TCF7L2 gene and found that three SNPs are associated with susceptibility to type 2 diabetes in the Spanish population.
Following the initial report [2], there have been a number of association studies in various ethnic groups [3].
Recently variants in the TCF7L2 gene also were reported to be associated with beta cell function [7, 8] and response to sulfonylureas in Europids [9].
In our study, the three SNPs were significantly associated with an increased risk of type 2 diabetes. These results are in agreement with those recently reported in other European populations [3]. According to meta-analyses by Florez [10] and by Cauchi et al. [11] TCF7L2 is the most reproducible susceptibility gene for type 2 diabetes in various ethnic groups. In all ethnicities, TCF7L2 showed strong and consistent association, with the odds of developing type 2 diabetes being increased by 40–60% for each allele inherited [11]. Thus, our study provides confirmation that variants in the TCF7L2 gene are associated with type 2 diabetes in the Spanish population.
Interestingly our results show that common variants in the TCF7L2 gene associated with increased risk for type 2 diabetes are also associated with an increased proinsulin:insulin ratio after an OGTT.
The conversion of proinsulin to insulin is a fundamental step in insulin synthesis by the beta cell. Proinsulin levels have been reported to predict the risk of type 2 diabetes independently of insulin levels [12]. Furthermore, an elevated proinsulin:insulin ratio has been observed in conditions with impaired beta cell function such as type 2 diabetes [13]. Loos et al. [14] found an association between the risk allele of rs7903146 in the TCF7L2 gene and increased fasting proinsulin but not with insulin levels, suggesting that, in addition to insulin secretion, variants in the TCF7L2 might be involved in insulin synthesis and processing.
A recently published study in the German population found an association between TCF7L2 variants and reduced proinsulin to insulin conversion [15]. This study also showed fasting and 30 min proinsulin:insulin ratios significantly increased in carriers of the risk alleles in the TCF7L2 gene.
The precise mechanisms by which this gene affects diabetes risk are not known, but it is thought that a reduced efficiency in beta cell proinsulin processing might be the mediating factor in the TCF7L2–type 2 diabetes association [14]. Moreover, impaired proinsulin processing might be due to the impaired glucagon-like peptide 1 action present in carriers of the risk alleles in the TCF7L2 gene.
Furthermore, a recent paper demonstrated that Wnt signalling regulates the proliferation of pancreatic beta cells in vitro and in vivo [16].
Recently the rs7903146 T allele was associated with type 2 diabetes and decreased BMI, whereas a haplotype carrying the rs7903146 C allele and rs10885406 A polymorphisms was associated with increased BMI in type 2 diabetes individuals [17]. However, other authors [18], in agreement with our results, showed that the rs7903146 T allele frequencies were similar in normoglycaemic non-obese and obese individuals, suggesting no direct effect on obesity prevalence.
The lack of an association with insulin secretion in this study might be due to the index used (HOMA-B). Another limitation of our study is the relative small number of patients and control individuals.
In conclusion, the findings of our current study support the hypothesis that variants in the TCF7L2 gene might contribute to genetic susceptibility to type 2 diabetes in Spain, as in other southern European populations. The association with an increased proinsulin:insulin ratio after an OGTT suggests that TCF7L2 might be involved in insulin synthesis and processing. Further studies are warranted to understand the mechanisms that regulate TCF7L2 expression and function for translational human investigation.
Abbreviations
- HOMA-B:
-
homeostasis model assessment of beta cell function
- HOMA-IR:
-
homeostasis model assessment of insulin resistance
- SNP:
-
single nucleotide polymorphism
References
Florez JC, Hirschhorn J, Altshuler D (2003) The inherited basis of diabetes mellitus: implications for the genetic analysis of complex traits. Annu Rev Genomics Hum Genet 4:257–291
Grant SF, Thorleifsson G, Reynisdottir I et al (2006) Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 38:320–323
Cauchi S, Froguel P (2008) TCF7L2 genetic defect and type 2 diabetes. Curr Diab Rep 8:149–155
Florez JC, Jablonski KA, Bayley N et al (2006) TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N Engl J Med 355:241–250
Saxena R, Gianniny L, Burtt NP et al (2006) Common single nucleotide polymorphisms in TCF7L2 are reproducibly associated with type 2 diabetes and reduce the insulin response to glucose in nondiabetic individuals. Diabetes 55:2890–2895
Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC (1985) Homeostasis model assessment: insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28:412–419
Schäfer SA, Tschritter O, Machicao F et al (2007) Impaired glucagon-like peptide-1-induced insulin secretion in carriers of transcription factor 7-like 2 (TCF7L2) gene polymorphisms. Diabetologia 50:2443–2450
Lyssenko V, Lupi R, Marchetti P et al (2007) Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest 117:2155–2163
Pearson ER, Donnelly LA, Kimber C et al (2007) Variation in TCF7L2 influences therapeutic response to sulfonylureas: a GoDARTs study. Diabetes 56:2178–2182
Florez JC (2007) The new type 2 diabetes gene TCF7L2. Curr Opin Clin Nutr Metab Care 10:391–396
Cauchi S, El Achhab Y, Choquet H et al (2007) TCF7L2 is reproducibly associated with type 2 diabetes in various ethnic groups: a global meta-analysis. J Mol Med 85:777–782
Zethelius B, Hales CN, Lithell HO, Berne C (2004) Insulin resistance, impaired early insulin response, and insulin propeptides as predictors of the development of type 2 diabetes: a population-based, 7-year follow-up study in 70-year-old men. Diabetes Care 27:1433–1438
Røder ME, Porte D Jr, Schwartz RS, Kahn SE (1998) Disproportionately elevated proinsulin levels reflect the degree of impaired B cell secretory capacity in patients with noninsulin-dependent diabetes mellitus. J Clin Endocrinol Metab 83:604–608
Loos RJ, Franks PW, Francis RW et al (2007) TCF7L2 polymorphisms modulate proinsulin levels and beta-cell function in a British Europid population. Diabetes 56:1943–1947
Kirchhoff K, Machicao F, Haupt A et al (2008) Polymorphisms in the TCF7L2, CDKAL1 and SLC30A8 genes are associated with impaired proinsulin conversion. Diabetologia 51:597–601
Rulifson IC, Karnik SK, Heiser PW et al (2007) Wnt signaling regulates pancreatic beta cell proliferation. Proc Natl Acad Sci USA 104:6247–6252
Helgason A, Pálsson S, Thorleifsson G et al (2007) Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet 39:218–225
Cauchi S, Choquet H, Gutiérrez-Aguilar R et al (2008) Effects of TCF7L2 polymorphisms on obesity in European populations. Obesity 16:476–482
Acknowledgements
We thank the members of the Segovia Insulin Resistance Study Group. This work was supported by grants FEDER 2FD1997-2309 from Fondo Europeo de Desarrollo Regional; FISS 03/1618 from Fondo de Investigaciones Sanitarias; from Red de Centros RCMN (C03/08); a grant from Instituto de Salud Carlos III-RETIC RD06 (RD06/0015/0012); CIBER de Diabetes y Enfermedades Metabólicas Asociadas is an ISCIII project, Spain. Partial support also came from Educational Grants from Eli Lilly, Spain, and Bayer Pharmaceutical, Spain. We also acknowledge financial support by an Educational Grant from Santander Central Hispano Bank (GSCH 5292564).
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
González-Sánchez, J.L., Martínez-Larrad, M.T., Zabena, C. et al. Association of variants of the TCF7L2 gene with increases in the risk of type 2 diabetes and the proinsulin:insulin ratio in the Spanish population. Diabetologia 51, 1993–1997 (2008). https://doi.org/10.1007/s00125-008-1129-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-008-1129-2