Abstract
Aims/hypothesis
We compared metabolic gene expression in adipose tissue and skeletal muscle from patients with type 2 diabetes and from well-matched healthy control subjects. We hypothesised that gene expression would be discordantly regulated when comparing the two groups. Our secondary aim was to determine the effect of Interleukin-6 (IL6) infusion on circulating adipokines and on gene expression in human adipose tissue. To do this we used real-time RT-PCR.
Methods
Both diabetic and control subjects underwent basal skeletal muscle and subcutaneous adipose tissue biopsies. A subset of these individuals underwent a 3-h infusion of recombinant human IL6 and had adipose tissue samples taken before and after infusion.
Results
The mRNA gene expression of suppressor of cytokine signalling (SOCS) 3, peroxisome proliferative activated receptor (PPAR) alpha/delta, PPAR gamma, coactivator 1, alpha (PPARGC1A), carnitine palmitoyltransferase 1B and solute carrier family 2 (facilitated glucose transporter), member 4 (formerly known as glucose transporter 4/GLUT4), was higher in adipose tissue, but lower in skeletal muscle of diabetic patients than in that of control subjects. In addition, uncoupling protein 1 (UCP1) gene was detected in the adipose tissue of some of the diabetic patients, but not in the control subjects. The following genes were increased by infusion of recombinant human IL6 in both groups: SOCS1/3, resistin, adiponectin, AMP-activated protein kinase-alpha-1 and PPARA. Plasma tumour necrosis factor alpha, adiponectin and resistin were all unaffected by IL6 infusion, but plasma resistin was lower in the diabetic subjects than in control subjects.
Conclusions/interpretation
The observation that PPARGC1A and the PPARs were upregulated in the adipose tissue of type 2 diabetic patients, along with the finding that adipose tissue from some patients with type 2 diabetes can express UCP1 mRNA, suggests that in these patients white adipose tissue may move towards a brown adipose tissue phenotype.
Similar content being viewed by others
Introduction
Skeletal muscle and adipose tissue are two of the most important metabolic tissues but they regulate metabolism differently. Adipocytes are a sink for fatty acids, whereas skeletal muscle primarily oxidises fatty acids. Drugs such as the thiazolidinediones can act by increasing fatty acid storage in adipose tissue while concomitantly reducing fat storage in skeletal muscle [1]. Skeletal muscle transports quantitatively more glucose for storage and oxidation than other tissues [2]. It is also well established that patients with type 2 diabetes have coordinated downregulation of genes that regulate metabolism, in particular those associated with oxidative phosphorylation in skeletal muscle [3, 4]. It is likely that this coordinated downregulation of metabolic gene expression is primarily the result of inactivity but it may also be influenced by nutrient excess and/or genetics [3]. When compared with skeletal muscle gene expression, there is little research into the effect of type 2 diabetes on gene expression in adipose tissue. Protection from insulin resistance induced by knockout of the adipocyte protein perilipin shows that adipocytes tend to upregulate oxidative pathways [5] in a similar fashion to human skeletal muscle [3, 4]. Few studies have examined the regulation of genes in adipose tissue and muscle from patients with type 2 diabetes. Genes associated with insulin action, such as hexokinase 2, phosphoinositide-3-kinase regulatory subunit 1 and sterol regulatory element binding transcription factor 1 c (SREBF1C), are coordinately downregulated in insulin-stimulated muscle and adipose tissue from patients with type 2 diabetes compared with that from control subjects [6]. Surprisingly, no studies have examined the expression of genes that are important in the control of metabolism in both muscle and adipose tissue in patients with type 2 diabetes compared with healthy control subjects. On the one hand, it may be expected that the metabolic gene expression profile would be coordinated when comparing gene expression in adipose tissue with that in skeletal muscle because factors that influence metabolism often circulate and act via transmembrane receptors to activate key transcription factors such as the peroxisome proliferative activated receptor-γ (PPARG), which is a target for insulin-sensitising therapies [7]. On the other hand, muscle contraction can initiate gene transcription within the cell as a result of factors such as elevated cytosolic calcium levels, factors that would have no effect on adipocyte gene transcription [8] and that lead to discordant gene expression when comparing the two tissue types. Indeed, in a recent study, mRNA gene expression of suppressor of cytokine signalling (SOCS) 3 was found to be discordantly regulated; it was reduced in skeletal muscle but increased in adipose tissue when comparing obese with lean subjects [9]. Since therapeutics for insulin resistance are now being developed in a tissue-specific manner [10], it is important to compare metabolic gene expression in insulin-sensitive tissue from patients with type 2 diabetes. Hence, this was the first aim of this study and we hypothesised that metabolic gene expression would be discordantly regulated in adipose tissue and skeletal muscle from patients with type 2 diabetes compared with those from healthy controls.
The effect of the cytokine IL6 on the aetiology of insulin sensitivity is the subject of debate. We recently showed that infusion of IL6 into patients with type 2 diabetes normalised insulin levels without affecting glucose clearance [11], raising the possibility that acute IL6 infusion may augment insulin sensitivity. In support of this notion, Weigert et al. [12, 13] recently demonstrated that insulin signalling and glycogen synthesis were increased by IL6 administration in L6 myotubes, while whole-body IL6 knockout renders mice obese and glucose intolerant [14]. In contrast, IL6 treatment impairs liver insulin signalling [15] and leads to impaired insulin action during an insulin tolerance test [16] and a euglycaemic–hyperinsulinaemic clamp [17] in mice. Although the literature is controversial, the common perception is that IL6 is an ‘adipokine’ that is elevated in obesity to impair insulin action [7] even though IL6 bears no relation to insulin action during a hyperinsulinaemic–euglycaemic clamp in humans [18]. To better understand the acute effects of IL6 on metabolism, a secondary aim of this study was to determine the effect of IL6 infusion on circulating adipokines and on gene expression in human adipose tissue.
Subjects and methods
Subjects
Tissue biopsies and fasting blood samples were obtained from 13 elderly male patients with type 2 diabetes (age: 57.5±4.1 years; weight: 90.2±5.2 kg; BMI: 28.4±1.1 kg/m2; fasting glucose: 8.8±0.8 mmol/l; fasting insulin: 104.8±17.1 pmol/l; fasting NEFA: 1.06±0.08 mmol/l) and 12 healthy male control subjects (age: 56.8±2.9 years; weight: 87.2±1.9 kg; BMI: 27.9±0.7 kg/m2; fasting glucose: 5.0±0.2 mmol/l; fasting insulin: 48.9±6.5 pmol/l; fasting NEFA: 0.78±0.04 mmol/l), who were statistically similar in age, weight and BMI. The experimental protocols were approved by the Human Ethics Committee of RMIT University and the Ethical Committee of Copenhagen and Fredriksberg Communities, Denmark, and were performed according to the ‘Declaration of Helsinki’. The categorisation of patients to the diabetes group was determined by a fasting blood glucose concentration >7.0 mmol/l, and >11.1 mmol/l following an oral glucose tolerance test, which is in accordance with the WHO 1999 criteria. None of the patients received insulin treatment, nor were any of them taking oral hypoglycaemic agents or any other medication known to alter carbohydrate metabolism. Subjects were informed of the possible risks and discomfort involved before giving their written consent to participate. The patients with type 2 diabetes had a mean time of 4±1 years since diagnosis (range 6 months to 10 years), and did not partake in any regular physical exercise. Other than diabetes, none of the subjects had any significant medical problems and all were non-smokers. Subjects were instructed to abstain from any form of vigorous physical activity for 36 h prior to an experiment (described subsequently), and to maintain their normal diet.
Experimental procedure
The subjects reported to the laboratory following a 12–14 h overnight fast and underwent muscle and fat biopsies. Under local anaesthesia (lidocaine), a subcutaneous adipose tissue sample was obtained from an incision made ∼4 cm lateral to the umbilicus and a muscle biopsy was obtained from the vastus lateralis from an incision made 4 cm proximal from the lateral epicondyle of the femur [18, 19]. A subset of subjects(sevenpatientswithtype2diabetes[67.4±3.6 years, 86.9±6.8 kg, BMI: 27.3±1.7 kg/m2] and six healthy male controls [63.8±3.4 years, 88.6±2.0 kg, BMI: 28.3±1.1 kg/m2]) visited the laboratory on two occasions, separated by 7–14 days. On one occasion, subjects were infused with recombinant human (rh) IL6, and on another occasion they were infused with isotonic saline. Pilot work and data from previous studies [11] demonstrated that IL6 delivered in 2% human albumin at a rate of 7 μg/h would result in a concentration of ∼140 pg/ml, which is seen during strenuous exercise. This dose was administered to all subjects during IL6 treatment. Subjects were randomly assigned and blinded to either the saline or the rhIL6 treatment, which was delivered via a peristaltic pump at a flow rate of 15 ml/h into an antecubital vein. IL6 or saline was infused for a period of 180 min and adipose tissue and venous blood samples were obtained before, immediately following and 2 h after the end of the infusion.
To determine the effect of IL6 infusion on circulating adipokines, ELISA was used to measure plasma levels of IL6, TNF, adiponectin (ADIPOQ) and resistin (RETN) [20, 21]. Fasting blood samples were also analysed for glucose, insulin and NEFA as previously described [11].
Candidate gene analysis using real-time RT-PCR
A portion of adipose tissue (50–75 mg) or skeletal muscle (20–30 mg) was extracted for total RNA using the modified acid guanidinium thiocyanate–phenolchloroform extraction of Chomczynski and Sacchi [22]. Samples were reverse transcribed using Taqman reverse transcription reagents (Applied Biosystems, Foster City, CA, USA) at a final concentration of 10 ng/μl. In the present study, we used a candidate gene approach and, therefore, we could not examine all the genes associated with metabolism. Accordingly, we chose to study the mRNA expression of genes that have been implicated in the aetiology of type 2 diabetes and the mRNA expression of genes encoding key secretory adipokines. We acknowledge that some key genes may have been missed using this approach. Nonetheless, we examined a relatively wide array of genes. PCR were completed with individual reactions of 5–10 μg total RNA, 1× BioRad iQ Supermix (BioRad, Hercules, CA, USA), 50–200 nM Taqman fluorescent probe (Applied Biosystems) and 300–900 nM PCR primers (Sigma Genosys, Castle Hill, NSW, Australia) in a total volume of 25 μl using a Bio-Rad iCycler (Bio Rad). Primers and Taqman probes were designed using Primer Express software (Applied Biosystems) to appropriate specifications for all genes except those encoding uncoupling protein 1 (UCP1) and ADIPOQ. For these two genes probe/primer kits were purchased from Applied Biosystems (TaqMan Gene Expression Assays; UCP1 Assay ID: Hs0022453_m1, GenBank NM 021833, ADIPOQ Assay ID: Hs00605917_m1, GenBank NM004797). Probes were designed with the 6-carboxyfluorescein reporter dye on the 5′ end and 6-carboxytetramethylrhodamine quencher dye to the 3′ end. The primer and probe sequences are listed in a table published as Electronic Supplementary Material (ESM, ESM Table 1). 18S mRNA was quantified using predeveloped reagents (Applied Biosystems) in the same reaction as the gene of interest for use as a constitutively expressed ‘house-keeping’ gene. To limit the possibility of nonspecific amplification of each gene, we designed the nucleic acid sequences to span an exon–exon junction when possible to limit the possibility of genomic DNA contamination.
We quantified gene expression using a multiplex comparative critical threshold (CT) method (iCycler iQ; Bio-Rad, Hercules, CA) as previously described [23]. A CT value reflects the cycle number at which DNA amplification is first detected. This method detected our reference gene (18S) and the gene of interest in a single tube, where the primers for 18S were limited to ensure that adequate amounts of reagents were present for the amplification of both genes. It was possible to detect 18S in the same tube as our genes of interest because the reporter dyes attached to the TaqMan probes fluoresce at different emission wavelength maxima. In preliminary experiments, we determined the relative efficiency of amplification of 18S versus the genes of interest. These experiments revealed approximately equal efficiencies of the amplification of 18S and the genes of interest over different starting template concentrations. We also performed experiments to demonstrate that multiplex versus nonmultiplex experiments had no effect on CT values, and that primer-limited multiplex 18S versus nonprimer-limited, nonmultiplex 18S reactions also had no effect on CT values. Finally, we have determined the linear dynamic range for starting template concentrations.
For each sample, a ΔCT value was obtained by subtracting 18S CT values from the CT of the gene of interest. To determine the basal mRNA expression when comparing groups, we used the value taken in control subjects as the control. Hence, a ΔCT value was obtained for each of the control subjects and averaged. This value was subtracted from the ΔCT value for each diabetic subject to derive a Δ–ΔCT value. The expression of each gene was then evaluated by 2−Δ–ΔC T. Because the ΔCT value for each non-diabetic subject was 0, the 2−Δ–ΔC T for this group was designated 1.
To determine the effect of IL6 infusion on the mRNA expression in each group, a ΔCT value was obtained by subtracting 18S CT values from each gene of interest, using the preinfusion value as the control. Preinfusion values for each subject were subtracted from the other samples for each subject to derive a 2−Δ–ΔC T value. The expression was then evaluated by 2−Δ–ΔC T, with all preinfusion values for each subject being 1.
Statistical analyses
All data are presented as mean±SEM. Group physical characteristics were analysed using a one-way analysis of variance (ANOVA). Newman–Kuel’s post-hoc tests were used where significant differences from ANOVA were revealed. Significance was accepted as p<0.05. All statistics were completed using Statistica software for windows (StatSoft, version 5.1, 1997, Statistica, Tulsa, OK, USA).
Results
The mRNA expression of metabolic genes is discordantly expressed in adipose tissue and skeletal muscle of patients with type 2 diabetes
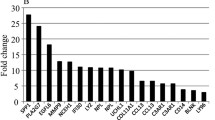
A comparison between the expression of selected genes in skeletal muscle and adipose tissue that are involved in the regulation of metabolism in these tissues is shown in Fig. 1. The expression of a number of genes was altered in patients with type 2 diabetes. In adipose tissue, the expression of SOCS3, peroxisome proliferator activated receptor-α (PPARA), peroxisome proliferator activated receptor-δ (PPARD), the gene encoding PPAR gamma coactivator 1 alpha (PPARGC1A) and carnitine palmitoyl transferase 1 (muscle isoform, CPT1B) was increased in diabetic subjects compared with controls, whereas expression of pyruvate dehydrogenase kinase 4 (PDK4) and uncoupling protein (UCP3) was reduced (p<0.05). In skeletal muscle IL6 and PDK4 were elevated in patients, whereas SOCS3, PPARA, PPARD, SLC2A4, CPT1B, fatty acid translocase (FAT/CD36), PPARGC1A, SOCS1, SREBF1A and SREBF1C were significantly reduced in diabetic patients compared with controls (p<0.05). As the expression of PPARA, PPARD, PPARGC1A and CPT1B was increased in the adipose tissue of diabetic subjects compared with controls, we hypothesised that human adipose tissue from patients with diabetes may be adopting a brown adipose tissue genotype because it has been shown that cultured human adipose tissue can express ‘brown fat cell features’ when transfected with PPARGC1A [24]. Accordingly, we examined our adipose tissue and muscle cDNA samples for UCP1 mRNA expression. As expected, we did not detect UCP1 in any muscle samples or in any adipose samples from control subjects. However, in ∼66% of patients with diabetes, we detected UCP1 mRNA expression in their adipose tissue, although no correlation was observed between PPARGC1A and UCP1 expression, possibly because not all the patients expressed UCP1. Since we used the CT method to express the data (described in detail above), it was not possible to quantify the UCP1 mRNA values in the patients who expressed this in their adipose tissue. However, the ΔCT for the patients who expressed UCP1 in their adipose tissue averaged 25.7±0.6 and ranged from 22 to 27, indicating that the level of expression between these patients was broad.
Basal expression of metabolic genes in skeletal muscle (closed bars) and adipose tissue (open bars) in patients with type 2 diabetes compared with age- and BMI-matched control subjects (set at 1, dashed line). * p<0.05 for difference in diabetic patients compared with controls (adipose tissue); # p<0.05 for difference in diabetic patients compared with controls (skeletal muscle). † mRNA not detectable. Values are expressed as mean±SE (diabetic subjects n=13, control subjects n=12)
Plasma adipokines are not affected by IL6 infusion but circulating resistin is lower in patients with type 2 diabetes
Circulating concentrations of cytokines are shown in Table 1. Concentrations of IL6 were not different between the control and diabetic subjects at any time points, but IL6 concentrations increased (p<0.01) in both groups as a result of IL6 infusion, and also during saline infusion (p<0.05). Although the concentrations attained as a result of infusion were higher than predicted (∼200 versus ∼140 pg/ml), subjects were not aware of the treatments, did not feel any ill effects of rhIL6 infusion and did not exhibit any increase in body temperature or heart rate compared with vehicle infusion (data not shown). Neither plasma TNF-α nor ADIPOQ was different when comparing diabetic patients with control subjects, nor were plasma concentrations of these cytokines affected by infusion. In contrast, while plasma resistin was unaffected by IL6 treatment, circulating levels were lower (p<0.01) in diabetic patients compared with control subjects (Table 1).
IL6 infusion increases adipose tissue expression of metabolic genes
Using real-time RT-PCR, none of the measured metabolic genes in adipose tissue responded differently to IL6 infusion in the diabetic patients compared with the control subjects. As a result the data presented in Fig. 2 represent pooled data from both populations. SOCS1 and SOCS3 mRNA expression were markedly increased by IL6 infusion. This was expected because the genes encoding SOCS proteins are activated by signal transducer and activator of transcription 1 and 3 proteins, which are phosphorylated via activation of the IL6R/gp130 receptor complex (for review see [25]). Immediately after infusion of IL6, RETN and AMP-activated protein kinase (PRKAA1) genes were elevated (p<0.05), while PPARG/D, TNF, and ADIPOQ showed a tendency (p<0.15) to be elevated. Three hours after infusion, SOCS3 and PRKAA1 remained elevated, while PPARA and ADIPOQ were higher (p<0.05) compared with saline infusion.
Candidate gene expression in adipose tissue from patients with type 2 diabetes and healthy control subjects immediately after (a) and 120 min after (b) 180 min of continuous infusion of vehicle (saline mixed in human albumin; open bars) or rhIL6 (mixed in albumin; closed bars). Data, which were pooled because no differences were observed between patients and control subjects, are expressed relative to preinfusion values (set at 1, dotted lines). * p<0.05 for difference between IL6 and vehicle. Values are expressed as mean±SE (n=13)
Discussion
It is well known that genes that regulate metabolism and oxidative phosphorylation are coordinately downregulated in the skeletal muscle of patients with type 2 diabetes [3, 4]. Consistent with these previous studies, we show that the expression of PPARGC1A, PPARA, PPARD, FAT/CD36 and CPT1B, genes that encode key proteins in oxidative phosphorylation, was downregulated in skeletal muscle of patients with type 2 diabetes. However, somewhat surprisingly, the mRNA expression of these genes was coordinately upregulated in the adipose tissue from these patients. This finding, coupled with the fact that we detected UCP1 in the adipose tissue of most of the patient population, but in none of the control subjects, suggests that in type 2 diabetes the white adipose tissue may be adopting a brown adipose tissue phenotype.
The observation that PPARGC1A mRNA was more than three-fold higher when comparing diabetic with control subjects was surprising because two previous studies have observed the opposite [26, 27]. However, given that we observed coordinate upregulation of metabolic genes and that these analyses were conducted concomitantly with the cDNA from the skeletal muscle samples, we are confident that our data accurately reflect mRNA expression in adipose tissue in vivo. To examine further the coordinate upregulation of metabolic genes in adipose tissue, we determined the level of expression of UCP1 in our samples. UCP1 is primarily expressed in brown adipose tissue and because brown adipose tissue disappears soon after birth in large mammals, the level of expression of UCP1 mRNA in white adipose tissue is extremely low if indeed present at all [28, 29]. We could not detect UCP1 mRNA in skeletal muscle, nor could we detect it in the adipose tissue of healthy humans by real-time RT-PCR. However, in the majority of the patients with type 2 diabetes, we were able to measure UCP1 mRNA abundance, and even though others have detected low expression levels of UCP1 mRNA in the extraperitoneal adipose tissue of healthy individuals [29], we did not detect this. Our observation of parallel upregulation of PPARGC1A and UCP1 mRNA in the adipose tissue of patients with type 2 diabetes is consistent with previous data showing that adenoviral-mediated overexpression of PPARGC1A in cultured white adipocytes upregulates UCP1 in vitro [24]. We cannot determine, from our data, why the adipose tissue of patients with type 2 diabetes would display this phenotype. However, because type 2 diabetes is often a disorder mediated by improper energy balance whereby lipid storage exceeds energy expenditure, it is tempting to speculate that the shift towards a brown adipose tissue phenotype represents a mechanism whereby increased utilisation of fat within white adipose tissue is an endogenous defence against progression of the disease. It should be noted, that the BMI for both the diabetic patients and the control group was ∼28 and, therefore, our subjects were overweight rather than obese. It is possible, therefore, that the pattern of gene expression in this subset of type 2 diabetics allowed them to be ‘obesity resistant’, such that fatty acids are oxidised, rather than stored, by the adipocyte. To further examine this possibility, we looked at the pattern of mRNA expression for both PPARGC1A and UCP1 in the adipose tissue of the diabetic subjects and performed correlations between level of expression (or detection for UCP1) and BMI. No significant correlations were observed in our patients, whose BMI ranged from 24 to 36.
One interesting finding from the present study was the observation that circulating RETN levels were markedly lower in patients with type 2 diabetes (Table 1). This observation seems anomalous because Lazar and colleagues [30, 31] have shown that the in vivo administration of recombinant RETN to mice causes insulin resistance, whereas administration of an anti-RETN antibody increased the insulin sensitivity of obese and insulin-resistant mice. Moreover, cultured adipocytes incubated with RETN are insulin resistant, a phenotype prevented by incubating cells with an anti-RETN antibody. Despite these findings in animals and in cell culture, the data with respect to humans are the subject of controversy [32]. In agreement with some [33, 34] but not all [35] previous investigations, we observed no difference in RETN mRNA expression in adipose tissue between diabetic and control subjects. Recently, Ng et al. [36] demonstrated that in the newborn offspring of mothers with type 2 diabetes, plasma RETN was markedly reduced in those infants who displayed markedly elevated basal glucose and insulin. Our data are consistent with these recent data. It must be acknowledged that our data do not necessarily question the association of RETN with insulin resistance because a reduction in circulating RETN may be a mechanism to protect the patient against further insulin resistance. Recently, another adipose-tissue-derived cytokine, namely visfatin, was found to be elevated in obese humans but was also found to bind and activate the insulin receptor [37]. Together, these data suggest that the adipose tissue may have the capacity to upregulate the release of insulin-sensitising agents or conversely downregulate the secretion of factors that exacerbate insulin resistance.
The effect of IL6 infusion on mRNA expression levels had a minimal effect on gene expression, although it must be noted that the infusion period was limited to 180 min. Of note, none of the measured genes was reduced by IL6 infusion. There appeared to be no consistency in the regulation of metabolic gene expression by IL6 infusion. Genes encoding proteins that have been linked to insulin resistance such as SOCS1/3, RETN and TNF were upregulated by IL6 but, conversely, genes that encode proteins normally associated with enhanced insulin sensitivity, namely PRKAA1, PPARD and ADIPOQ, were also increased, making it difficult for conclusions to be drawn.
One limitation of our study was that we studied our subjects only in the fasted state and it would have been ideal to compare the gene expression in different tissues between different groups in both the fed and fasted states. Although the differences observed between the patient and control populations are likely to be the result of population differences, and not of the effect of fasting, we cannot rule out the possibility that the patient population responded to an overnight fast differently when compared with the control group. For example, the differences in fasting NEFA levels may have been blunted in the fed state, which could potentially have influenced the genes that encode proteins targeted by fatty acids, e.g. the PPARs.
In conclusion, we demonstrated that the genes that regulate oxidative phosphorylation are discordantly regulated in adipose tissue and skeletal muscle from patients with type 2 diabetes. The observation that PPARGC1A mRNA was approximately three-fold higher in the adipose tissue of diabetic patients than in that of healthy controls, coupled with the detection of UCP1 in the adipose tissue of most of the patients, whereas it was undetected in the healthy subjects, suggests that in type 2 diabetes the adipose tissue may be shifting toward a brown adipose tissue phenotype. We also conclude that, despite the association of the adipokine RETN with insulin resistance, plasma concentrations of RETN are markedly lower in patients with type 2 diabetes.
Abbreviations
- ADIPOQ:
-
adiponectin
- CPT1B:
-
carnitine palmitoyl transferase 1 (muscle isoform)
- FAT/CD36:
-
fatty acid translocase
- IL6:
-
interleukin-6
- PDK4:
-
pyruvate dehydrogenase kinase 4
- PRKA:
-
AMP-activated protein kinase
- PPAR:
-
peroxisome proliferative activated receptor
- PPARGC1A:
-
peroxisome proliferative activated receptor gamma coactivator 1 alpha
- RETN:
-
resistin
- SLC2A4:
-
solute carrier 2A (facilitated glucose transporter)
- SOCS:
-
suppressor of cytokine signalling
- SREBF1:
-
sterol regulatory element binding factor
- UCP:
-
uncoupling protein
References
Ye JM, Dzamko N, Cleasby ME et al (2004) Direct demonstration of lipid sequestration as a mechanism by which rosiglitazone prevents fatty-acid-induced insulin resistance in the rat: comparison with metformin. Diabetologia 47:1306–1313
Zurlo F, Larson K, Bogardus C, Ravussin E (1990) Skeletal muscle metabolism is a major determinant of resting energy expenditure. J Clin Invest 86:1423–1427
Patti ME, Butte AJ, Crunkhorn S et al (2003) Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A 100:8466–8471
Mootha VK, Lindgren CM, Eriksson KF et al (2003) PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34:267–273
Castro-Chavez F, Yechoor VK, Saha PK et al (2003) Coordinated upregulation of oxidative pathways and downregulation of lipid biosynthesis underlie obesity resistance in perilipin knockout mice: a microarray gene expression profile. Diabetes 52:2666–2674
Ducluzeau PH, Perretti N, Laville M et al (2001) Regulation by insulin of gene expression in human skeletal muscle and adipose tissue. Evidence for specific defects in type 2 diabetes. Diabetes 50:1134–1142
Lazar M (2005) How obesity causes diabetes: not a tall tale. Science 307:373–375
Febbraio MA, Pedersen BK (2002). Muscle-derived interleukin-6: mechanisms for activation and possible biological roles. FASEB J 16:1335–1347
Rieusset J, Bouzakri K, Chevillotte E et al (2004) Suppressor of cytokine signaling 3 expression and insulin resistance in skeletal muscle of obese and type 2 diabetic patients. Diabetes 53:2232–2241
Masuzaki H, Flier JS (2003) Tissue-specific glucocorticoid reactivating enzyme, 11 beta-hydroxysteroid dehydrogenase type 1 (11 beta-HSD1)—a promising drug target for the treatment of metabolic syndrome. Curr Drug Targets Immune Endocr Metabol Disord 3:255–262
Petersen EW, Carey AL, Sacchetti M et al (2005) Acute IL-6 treatment increases fatty acid turnover in elderly humans in vivo and in tissue culture in vitro. Am J Physiol Endocrinol Metab 288:E155–E162
Weigert C, Brodbeck K, Staiger H et al (2004) Palmitate, but not unsaturated fatty acids, induces the expression of interleukin-6 in human myotubes through proteasome-dependent activation of nuclear factor-kappaB. J Biol Chem 279:23942–23952
Weigert C, Hennige AM, Brodbeck K, Haring HU, Schleicher ED (2005). Interleukin-6 (IL-6) acts as insulin sensitizer on glycogen synthesis in human skeletal muscle cells by phosphorylation of Ser-473 of Akt. Am J Physiol Endocrinol Metab 289:E251–E257
Wallenius V, Wallenius K, Ahren B et al (2002). Interleukin-6-deficient mice develop mature-onset obesity. Nat Med 8:75–79
Senn JJ, Klover PJ, Nowak IA, Mooney RA (2002) Interleukin-6 induces cellular insulin resistance in hepatocytes. Diabetes 51:3391–3399
Klover PJ, Zimmers TA, Koniaris LG, Mooney RA (2003) Chronic exposure to interleukin-6 causes hepatic insulin resistance in mice. Diabetes 52:2784–2789
Kim HJ, Higashimori T, Park SY et al (2004) Differential effects of interleukin-6 and -10 on skeletal muscle and liver insulin action in vivo. Diabetes 53:1060–1067
Carey AL, Bruce CR, Sacchetti M et al (2004) Interleukin-6 and tumor necrosis factor-alpha are not increased in patients with type 2 diabetes: evidence that plasma interleukin-6 is related to fat mass and not insulin responsiveness. Diabetologia 47:1029–1037
Plomgaard P, Keller P, Keller C, Pedersen BK (2005) TNF-a, but not IL-6, stimulates plasminogen activator inhibitor 1 (PAI-1) expression in human subcutaneous adipose tissue. J Appl Physiol 98:2019–2023
Steensberg A, Keller C, Starkie RL, Osada T, Febbraio MA, Pedersen BK (2002) IL-6 and TNF-alpha expression in, and release from, contracting human skeletal muscle. Am J Physiol Endocrinol Metab 283:E1272–1278
Lindegaard B, Keller P, Bruunsgaard H, Gerstoft J, Pedersen BK (2004) Low plasma level of adiponectin is associated with stavudine treatment and lipodystrophy in HIV-infected patients. Clin Exp Immunol 135:273–279
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159
Bruce CR, Carey AL, Hawley JA, Febbraio MA (2003) Intramuscular heat shock protein 72 and heme oxygenase-1 mRNA are reduced in patients with type 2 diabetes: evidence that insulin resistance is associated with a disturbed antioxidant defense mechanism. Diabetes 52:2338–2345
Tiraby C, Tavernier G, Lefort C et al (2003) Acquirement of brown fat cell features by human white adipocytes. J Biol Chem 278:33370–33376
Carey AL, Febbraio MA (2004) Interleukin-6 and insulin sensitivity: friend or foe? Diabetologia 47:1135–1142
Semple RK, Crowley VC, Sewter CP et al (2004) Expression of the thermogenic nuclear hormone receptor coactivator PGC-1alpha is reduced in the adipose tissue of morbidly obese subjects. Int J Obes Relat Metab Disord 28:176–179
Hammarstedt A, Jansson PA, Wesslau C, Yang X, Smith U (2003) Reduced expression of PGC-1 and insulin-signaling molecules in adipose tissue is associated with insulin resistance. Biochem Biophys Res Commun 301:578–582
Krief S, Lonnqvist F, Raimbault S et al (1993) Tissue distribution of beta 3-adrenergic receptor mRNA in man. J Clin Invest 91:344–349
Oberkofler H, Dallinger G, Liu YM, Hell E, Krempler F, Patsch W (1997) Uncoupling protein gene: quantification of expression levels in adipose tissues of obese and non-obese humans. J Lipid Res 38:2125–2133
Steppan CM, Bailey ST, Bhat S et al (2001) The hormone resistin links obesity to diabetes. Nature 409:307–312
Steppan CM, Brown EJ, Wright CM et al (2001) A family of tissue-specific resistin-like molecules. Proc Natl Acad Sci U S A 98:502–506
Hotamisligil GS (2003) The irresistible biology of resistin. J Clin Invest 111:173–174
Nagaev I, Smith U (2001) Insulin resistance and type 2 diabetes are not related to resistin expression in human fat cells or skeletal muscle. Biochem Biophys Res Commun 285:561–564
Janke J, Engeli S, Gorzelniak K, Luft FC, Sharma AM (2002) Resistin gene expression in human adipocytes is not related to insulin resistance. Obes Res 10:1–5
Savage DB, Sewter CP, Klenk ES et al (2001) Resistin/Fizz3 expression in relation to obesity and peroxisome proliferator-activated receptor-gamma action in humans. Diabetes 50:2199–2202
Ng PC, Lee CH, Lam CW, Wong E, Chan IH, Fok TF (2004) Plasma ghrelin and resistin concentrations are suppressed in infants of insulin-dependent diabetic mothers. J Clin Endocrinol Metab 89:5563–5568
Fukuhara A, Matsuda M, Nishizawa M et al (2005) Visfatin: a protein secreted by visceral fat that mimics the effects of insulin. Science 307:426–430
Acknowledgements
We acknowledge the subjects for their participation. In addition, the technical support of R. Rousing, H. Willumsen and C. Nielsen is greatly appreciated. This work was supported by The National Health and Medical Research Council of Australia (Grant-251558), The Danish National Research Foundation (Grant 504-14), and The Danish Medical Research Council (Grant 22-01-009). M. A. Febbraio is supported by a Senior Research Fellowship from The National Health and Medical Research Council of Australia.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Carey, A.L., Petersen, E.W., Bruce, C.R. et al. Discordant gene expression in skeletal muscle and adipose tissue of patients with type 2 diabetes: effect of interleukin-6 infusion. Diabetologia 49, 1000–1007 (2006). https://doi.org/10.1007/s00125-006-0178-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-006-0178-7