Abstract
Aims/hypothesis
Although genetic susceptibility plays an important role in the pathogenesis of type 2 diabetes, most of the genes that influence susceptibility to type 2 diabetes have yet to be identified. Krüppel-like transcription factors are known to play important roles in development and cell differentiation, and have recently been implicated in the pathogenesis of type 2 diabetes. The present study aimed to examine the associations of single nucleotide polymorphisms (SNPs) in genes encoding members of the Krüppel-like-factor (KLF) family with type 2 diabetes in a large cohort of Japanese subjects.
Methods
We genotyped 33 SNP loci found in 12 KLF genes in subjects with type 2 diabetes and in subjects from the general population using the PCR-Invader assay. We also examined the effects of the overexpression of KLF7 on adipogenesis in 3T3-L1 cells.
Results
We identified a significant association between an SNP in KLF7 and type 2 diabetes (A vs C: p=0.004 after Bonferroni’s correction, odds ratio=1.59, 95% CI 1.27–2.00). The expression of Klf7 decreased in response to the differentiation of 3T3-L1 adipocytes, and the overexpression of KLF7 resulted in significant inhibition of adipogenesis in 3T3-L1 cells.
Conclusions/interpretation
These results indicate that the gene encoding KLF7 is a novel candidate for conferring genetic susceptibility to type 2 diabetes.
Similar content being viewed by others
Introduction
Type 2 diabetes affects more than 100 million individuals worldwide [1]. The pathogenesis of this disease appears to involve insulin resistance in peripheral tissues combined with dysfunction of beta cells in pancreatic islets; however, the precise mechanism remains unknown [2, 3].
It is generally accepted that genetic factors contribute to the onset and progression of diabetes, and several genes responsible for specific forms of the disease, such as MODY and mitochondrial diabetes, have been identified [4, 5]. However, genetic alterations associated with these specific forms of diabetes account for only a small percentage of cases, and genes conferring susceptibility to type 2 diabetes have not been identified in the majority of patients.
The Krüppel-like factor (KLF) family of transcription factors was initially reported to play a pivotal role in cellular development and differentiation [6, 7]. Recently, members of the KLF family have been found to be involved in adipogenesis [8] and the expression of GLUT4 in adipocytes [9]. Because of this new role in fat metabolism, the KLF genes are thought to be candidates for conferring susceptibility to type 2 diabetes. However, to date, no genetic association studies have focused on the KLF genes.
In the present study, we present the results of a case–control study of several members of the KLF gene family in Japanese subjects, and provide evidence that KLF7 is a good candidate for conferring susceptibility to type 2 diabetes.
Subjects and methods
Subjects and DNA preparations
DNA samples were obtained from 1,130 patients with type 2 diabetes who regularly attended outpatient clinics at the Shiga University of Medical Science, the Tokyo Women’s Medical University, Juntendo University School of Medicine, Kawasaki Medical School, the Kawai Clinic, Toride Kyodo Hospital, Iwate Medical University School of Medicine, and Osaka City General Hospital (672 men, 458 women; age 60.1±11.6 years; duration of diabetes 13.5±10.0 years; HbA1c 7.2±1.2%; fasting plasma glucose 8.3±2.5 mmol/l; BMI 23.2±3.4 kg/m2 [all values are means±SD]). Diabetes was diagnosed according to the criteria of the World Health Organization. Type 2 diabetes was clinically defined as a gradual, adult onset of the disease. Subjects who tested positive for anti-glutamic acid decarboxylase antibody, and patients with mitochondrial disease (mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes [MELAS]) or MODY were excluded. The control group comprised 564 members of the general population who were recruited through several medical institutes in Japan (272 men, 292 women; age 48.3±23 years, BMI 24.9±3.6 kg/m2). We repeated the study in a second set of diabetic subjects (n=560; 349 men, 211 women; age 61.2±10.8 years; duration of diabetes 17.2±9.6 years; HbA1c 7.9±4.2%; fasting plasma glucose 8.4±2.9 mmol/l; BMI 24.6±3.7 kg/m2) and control subjects (n=359; 193 men 166 women; age 42.7±9.6 years). Written informed consent was obtained from each patient, and DNA extraction was performed using a standard phenol–chloroform procedure.
Genotyping
The single nucleotide polymorphisms (SNPs) that were used for genotyping experiments were selected from the IMS-JST Japanese database of SNPs (http://snp.ims.u-tokyo.ac.jp, last accessed in April 2005) [10, 11]. The genotype at each SNP locus was analysed using the Invader assay (Third Wave Technologies, Madison, WI, USA) or Taqman assay (Applied Biosystems, Foster City, CA, USA), as previously described [12]. The success rates of these assays were >95%, and there was almost 100% agreement between the results of genotyping and the results of direct sequencing [12]. Differences in the genotype and/or allele frequencies between the case group and the control group were analysed using 2×3 or 2×2 contingency tables. The protocol was approved by the Ethics Committee of the Institute of Physical and Chemical Research (Kanagawa, Japan).
Northern blot analysis
Mouse 3T3-L1 cells were grown to confluence and induced to differentiate into adipocytes using methods described previously [13]. Total RNA was extracted from the 3T3-L1 cells by Trizol reagent (Invitrogen, Carlsbad, CA, USA) at 0, 3 and 5 days after inducing adipocyte differentiation. Total RNA (20 μg) was separated by 1% denaturing formaldehyde agarose gel electrophoresis, and transferred onto a Hybond-N membrane (Amersham Biosciences, Piscataway, NJ, USA). Hybridisation was carried out using α-32P-labelled complementary DNA (cDNA) probes for mouse Klf7 and Gapd. Mouse cDNAs for Klf7 and Gapd were cloned by RT-PCR using the following primers: Klf7 sense, 5′-CCA CTG GAT GTC TGC CTT CT-3′, antisense, 5′-CAC TCG CAT CCT TCC CAT GA-3′; Gapd sense, 5′-CTT CTG CCG ATG CCC CCA T-3′, antisense, 5′- GCC TGC TTC ACC ACC TTC TTG AT-3′.
Construction of adenovirus vector and viral infection
An adenovirus vector encoding human KLF7 was prepared using the Adenovirus Expression Vector Kit (TaKaRa, Japan). Forty-eight hours before the induction of differentiation, 3T3-L1 cells were transduced with 100 plaque-forming units per cell for 12 h, and samples were obtained at the indicated days after inducing differentiation.
Oil red O staining
At the indicated days after inducing differentiation, the cells were washed twice with PBS, and then fixed for 2 h with 3.7% formaldehyde. Fixed cells were incubated with oil red O for 15 min at room temperature. After washing the cells four times with water, the stained lipid droplets in the cells were visualised by light microscopy. For quantification, the dye was extracted with isopropyl alcohol, and the absorbance was measured at 540 nm.
Statistical analysis
Statistical methods for determining the genotype–phenotype association and the Hardy–Weinberg equilibrium [14], and to calculate the linkage disequilibrium (LD) coefficients (D′ and Δ) have been described previously [15]. Differences between the case and control groups in terms of genotype distribution or allele frequency were analysed by the chi square test using 2×3 or 2×2 contingency tables, and p values below 0.05 were considered significant.
Results
We first genotyped 33 SNP loci within 12 KLF genes in 188 Japanese patients with type 2 diabetes and compared the allelic frequencies at these loci with those in the general population (n=564). Of the 33 SNP loci tested, only two exhibited a p value of less than 0.05 (Table 1). Both loci were found in KLF7 on chromosome 2q32. We then analysed 36 SNP loci within KLF7 using the maximum number of patients (n=1,130; see Table 1 of the Electronic supplementary material for details of these 36 SNPs). Several of these SNPs were found to be significantly associated with type 2 diabetes. In particular, we found that one of the alleles at an SNP locus in the second intron of KLF7 was strongly associated with type 2 diabetes (A vs C: p=0.004 after Bonferroni’s correction, odds ratio=1.59, 95% CI 1.27–2.00; Table 2). Because sex appeared to be distributed differently between the case and control groups, we subdivided the case group into male and female groups, and compared the genotype distribution for these two groups. The results indicated that the KLF7 genotype was not distributed differently between the male and female group (male: AA=82.8%, AC=16.5%, CC=0.6%; female: AA=83.2%, AC=15.9%, CC=0.9%), and a similar association between KLF7 and type 2 diabetes was observed in the two groups.
To exclude the possibility that the result obtained reflected associations between type 2 diabetes and other genes located near KLF7, we performed LD mapping of this region. This process identified a region of approximately 80 kb (20 kb upstream and 60 kb downstream of the landmark SNP; Fig. 1). We therefore concluded that the critical region for susceptibility to type 2 diabetes must lie within this 80-kb region, which did not contain any genes other than KLF7, indicating that KLF7 is the candidate gene.
We next examined the haplotype structure of the region surrounding KLF7. The analysis was performed by estimating haplotype phasing using the Expectation Maximisation (EM) algorithm [16] and by constructing haplotype blocks as previously described [17, 18]. The results indicated that 16 SNPs in KLF7 constitute one haplotype block with an allelic frequency of greater than 0.05, and that four common haplotypes define more than 90% of the population (Fig. 2). Subsequent examinations of the association of each haplotype with type 2 diabetes identified a significant relationship between haplotype 3 and type 2 diabetes; however, this association was not stronger than that found between the single locus (intron 2 +35092) and type 2 diabetes. Thus, the SNP in intron 2 (+35092, A/C) appeared to be able to explain the majority of the positive association between KLF7 and type 2 diabetes. We confirmed the association of this SNP with type 2 diabetes in an independent set of case and control subjects (Table 3).
Analysis of the haplotype structure and the estimated haplotype frequencies in KLF7. Sixteen SNPs, including the SNP at intron 2 +35092, constituted one haplotype block. The asterisk indicates the SNP at intron 2 +35092 (A/C). The p values for the comparison of haplotype frequencies between the case and control groups were generated by the chi square test using a 2×2 contingency table composed of the number of one haplotype and the sum of the other haplotypes
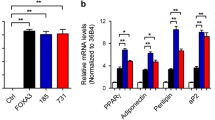
To establish whether this gene is involved in the pathogenesis of type 2 diabetes, we examined the pattern of expression of Klf7 in mouse 3T3-L1 cells at different stages of differentiation. As shown in Fig. 3, the expression of Klf7 was most abundant before inducing differentiation, and rapidly decreased after inducing differentiation. Furthermore, the overexpression of KLF7 in preadipocytes resulted in the remarkable inhibition of adipogenesis in 3T3-L1 cells (Fig. 3b–d).
a Northern blot analysis of mouse Klf7 using total RNA isolated from 3T3-L1 cells at 0, 3 and 5 days after the induction of differentiation. One transcript (8.3 kb, indicated by an arrow) was identified. The same blot was re-hybridised with Gapd used as a loading control. The blot shown is representative of the results obtained from three independent experiments. b Effect of KLF7 overexpression on adipogenesis. Cells were induced to differentiate into adipocytes 48 h after adenovirus infection. Oil red O staining was performed at the indicated days after inducing differentiation. − Uninfected cells; LacZ cells infected with adenovirus vectors encoding LacZ (control); KLF7 cells infected with adenovirus vectors encoding KLF7.c Oil red O staining of the cells at 10 days after the induction of differentiation (magnification ×100). The results shown are representative of two independent experiments. d Quantification of oil red O staining in uninfected cells (white bars), LacZ-infected cells (grey bars) and KLF7-infected cells (black bars)
Discussion
Although the contribution of genetic factors to the pathogenesis of type 2 diabetes is widely acknowledged, to date, only a few genes have been implicated in susceptibility to type 2 diabetes [19–21]. The difficulty involved in identifying alleles responsible for common diseases can be explained by the fact that the effects of individual genes in a complex genetic and environmental background are often too small to be identified. Therefore, a large-scale genotyping strategy, as employed in this study, can be more effective than a single candidate gene approach for the identification of susceptibility genes for common diseases such as diabetes mellitus. Because we examined total of 64 SNP loci, type 1 error due to multiple testing must be taken into account. We therefore corrected the p value for the association between a particular SNP in KLF7 (intron 2 +35092) and type 2 diabetes by Bonferroni’s method, and obtained a p value of approximately 0.004. Based on this, we concluded that the association between this SNP and type 2 diabetes was statistically significant. Because we used individuals from the general population as a control group, it is possible that the potential power of our study to detect an association of a particular locus with the disease was relatively weaker than if we had used subjects with normal glucose tolerance as a control group. Therefore, we examined the relationship between the SNP in KLF7 with type 2 diabetes in an independent set of case and control subjects to verify the association. Because the KLF7 locus, chromosome 2q32, was also reported to be a diabetes or relevant metabolic trait locus [22], we concluded that KLF7 was a good candidate gene for conferring susceptibility to type 2 diabetes.
KLF7 was originally identified by a degenerative PCR approach using a cDNA library from human vascular endothelial cells. Because the expression of KLF7 was detectable in wide variety of human tissues, it was then named ubiquitous Krüppel-like factor [23]. KLF7 was also identified as a transcriptional activator and was shown to be capable of increasing the expression of p21 WAF1/Cip1, which encodes a cell cycle regulator [24]. Furthermore, the gene encoding KLF7 is known to be predominantly expressed in the brain and the spinal cord in humans [23], and in the central and peripheral nervous system during embryogenesis in mice [24]. These findings suggest that KLF7 plays a pivotal role in the regulation of neurogenesis; however, the mechanism of its involvement in the pathogenesis of type 2 diabetes is not known.
In the present paper we have demonstrated that Klf7 expression can be detected in 3T3-L1 preadipocytes, and that expression decreases in response to the differentiation of 3T3-L1 adipocytes (Fig. 3a). Using cells infected with adenoviral vectors encoding KLF7, we have also demonstrated that KLF7 expression has an inhibitory effect on adipogenesis. This pattern of expression and effect on adipogenesis is similar to that previously reported for KLF2 [8], suggesting that KLF7 could also play a role in the regulation of adipogenesis. Therefore, it can be hypothesised that KLF7 may confer susceptibility to type 2 diabetes by affecting adipocyte function.
The exact mechanism by which the polymorphisms in KLF7 affect susceptibility to type 2 diabetes remains to be elucidated. By searching for additional polymorphisms within exons and the regulatory region of KLF7, we identified several additional polymorphisms within the gene (one synonymous SNP within exon 2, three SNPs within the 3′ untranslated region of exon 4, and three SNPs within 2 kb upstream of the transcription initiation site). However, a subsequent association study revealed that none of these SNPs was significantly associated with type 2 diabetes (data not shown).
In conclusion, we have identified KLF7 as a new candidate gene with specific alleles that can confer susceptibility to type 2 diabetes. Our results also suggest that KLF7 may contribute to the pathogenesis of type 2 diabetes by regulating the function of adipocytes.
Abbreviations
- KLF:
-
Krüppel-like factor
- LD:
-
linkage disequilibrium
- SNP:
-
single nucleotide polymorphism
References
Zimmet P, Alberti KGMM, Shaw J (2001) Global and societal implications of the diabetes epidemic. Nature 414:782–786
Kahn BB (1998) Type 2 diabetes: when insulin secretion fails to compensate for insulin resistance. Cell 92:593–596
Saltiel AR (2001) New perspectives into the molecular pathogenesis and treatment of type 2 diabetes. Cell 104:517–529
Fajans SS, Bell GI, Polonsky KS (2001) Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. N Engl J Med 345:971–980
Kadowaki T, Kadowaki H, Mori Y et al (1994) A subtype of diabetes mellitus associated with a mutation of mitochondrial DNA. N Engl J Med 330:962–968
Black AR, Black JD, Azizkhan-Clifford J (2001) Sp1 and Kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol 188:143–160
Bieker JJ (2001) Kruppel-like factors: three fingers in many pies. J Biol Chem 276:34355–34358
Banerjee SS, Feinberg MW, Watanabe M et al (2003) The Kruppel-like factor KLF2 inhibits peroxisome proliferator-activated receptor-gamma expression and adipogenesis. J Biol Chem 278:2581–2584
Gray S, Feinberg MW, Hull S et al (2002) The Kruppel-like factor KLF15 regulates the insulin-sensitive glucose transporter GLUT4. J Biol Chem 277:34322–34328
Hirakawa M, Tanaka T, Hashimoto Y, Kuroda M, Takagi T, Nakamura Y (2002) JSNP: a database of common gene variations in the Japanese population. Nucleic Acids Res 30:158–162
Haga H, Yamada R, Ohnishi Y, Nakamura Y, Tanaka T (2002) Gene-based SNP discovery as part of the Japanese Millennium Genome Project: identification of 190,562 genetic variations in the human genome. Single-nucleotide polymorphism. J Hum Genet 47:605–610
Ohnishi Y, Tanaka T, Ozaki K, Yamada R, Suzuki H, Nakamura Y (2001) A high-throughput SNP typing system for genome-wide association studies. J Hum Genet 46:471–477
Kishida K, Shimomura I, Kondo H et al (2001) Genomic structure and insulin-mediated repression of the aquaporin adipose (AQPap), adipose-specific glycerol channel. J Biol Chem 28:36251–36260
Nielsen DM, Ehm MG, Weir BS (1998) Detecting marker-disease association by testing for Hardy–Weinberg disequilibrium at a marker locus. Am J Hum Genet 63:1531–1540
Devlin B, Risch N (1995) A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics 29:311–322
Excoffier L, Slatkin M (1995) Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol 12:921–927
Gabriel SB, Schaffner SF, Nguyen H et al (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229
Tanaka N, Babazono T, Saito S et al (2003) Association of solute carrier family 12 (sodium/chloride) member 3 with diabetic nephropathy, identified by genome-wide analyses of single nucleotide polymorphisms. Diabetes 52:2848–2853
Horikawa Y, Oda N, Cox NJ et al (2000) Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet 26:163–175
Ong KK, Phillips DI, Fall C et al (1999) The insulin gene VNTR, type 2 diabetes and birth weight. Nat Genet 21:262–263
Deeb SS, Fajas L, Nemoto M et al (1998) A Pro12Ala substitution in PPAR2 associated with decreased receptor activity, lower body mass index, and improved insulin sensitivity. Nat Genet 20:284–287
Iwasaki N, Cox NJ, Wang YQ et al (2003) Mapping genes influencing type 2 diabetes risk and BMI in Japanese subjects. Diabetes 52:209–213
Matsumoto N, Laub F, Aldabe R et al (1998) Cloning the cDNA for a new human zinc finger protein defines a group of closely related Kruppel-like transcription factors. J Biol Chem 273:28229–28237
Laub F, Aldabe R, Friedrich V Jr, Ohnishi S, Yoshida T, Ramirez F (2001) Developmental expression of mouse Kruppel-like transcription factor KLF7 suggests a potential role in neurogenesis. Dev Biol 233:305–318
Acknowledgements
The authors would like to acknowledge S. Tsukada, K. Ohashi and the technical staff at the SNP Research Centre (Institute of Physical and Chemical Research, Kanagawa, Japan). This work was supported by a grant from the Japanese Millennium Project.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Kanazawa, A., Kawamura, Y., Sekine, A. et al. Single nucleotide polymorphisms in the gene encoding Krüppel-like factor 7 are associated with type 2 diabetes. Diabetologia 48, 1315–1322 (2005). https://doi.org/10.1007/s00125-005-1797-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-005-1797-0