Abstract
Aims/hypothesis
The SMXA-5 mouse is one of the SMXA recombinant inbred substrains established from the non-diabetic SM/J and A/J strains, and is a model for polygenic type 2 diabetes, characterised by moderately impaired glucose tolerance and hyperinsulinaemia. These diabetic traits are worsened by feeding a high-fat diet. The aim of this study was to dissect the diabetogenic loci in the A/J regions of the SMXA-5 genome that contribute to diabetes-related traits.
Materials and methods
We analysed the quantitative trait loci (QTL) for diabetes-related traits and obesity in (SM/J×SMXA-5)F2 intercross mice fed a high-fat diet. To verify the function of the responsible locus that was mapped in the present study, we constructed a congenic strain and characterised its diabetes-related traits.
Results
A major QTL for glucose tolerance, free-fed blood glucose concentration and BMI was mapped on chromosome 2. This locus existed near D2Mit15, with the highest logarithm of the odds score (12.6) for glucose concentration at 120 min in a glucose tolerance test, and was designated T2dm2sa. The diabetogenic allele of T2dm2sa originated in the A/J strain. SM.A-T2dm2sa, a congenic strain that introgressed the T2dm2sa region of A/J genome into SM/J, exhibited overt impaired glucose tolerance and hyperinsulinaemia.
Conclusions/interpretation
The development of impaired glucose tolerance in SM.A-T2dm2sa mice confirmed the results of QTL analysis for diabetes-related traits in F2 intercross mice. The present results suggest that there are latent diabetogenic loci in the genomes of non-diabetic A/J and SM/J mice, and that the coexistence of these loci, including T2dm2sa, causes impaired glucose tolerance in SMXA-5 and SM.A-T2dm2sa mice.
Similar content being viewed by others
Introduction
The development of type 2 diabetes is controlled by the interaction of multiple genetic and environmental factors [1]. The heterogeneity of the disease, which is a consequence of such interaction, makes it difficult to identify the genes involved in human diabetes. Environmental factors, such as typical Western diets, in which more than 50% of the calories are derived from fat, increase the risk of developing type 2 diabetes. As an environmental factor, dietary fat is a key component that influences metabolic pathways involved in the development of diabetes. Since genetic and environmental factors can be strictly controlled when inbred animal models are used, such models are invaluable for the dissection of complex diseases. Loci contributing to type 2 diabetes and related traits in diabetic animal models have been reported using quantitative trait locus (QTL) mapping analyses (GK rat [2, 3], OLETF rat [2, 4–7], SDT rat [8], NSY mouse [9], NZO mouse [10, 11], TSOD mouse [12], KK mouse [13], KK-Ay mouse [14, 15] and TH mouse [16]).
The SMXA-5 mouse is one of the 26 SMXA recombinant inbred (RI) substrains that have been established from parental strains SM/J and A/J [17]. Each SMXA RI strain has a mosaic genome derived from the SM/J and A/J strains. Although the parental strains were non-diabetic when fed a high-carbohydrate diet, SMXA-5 mice showed impaired glucose tolerance and hyperglycaemia [18, 19]. These results suggested that diabetogenic loci in the non-diabetic parental SM/J and A/J genomes coexist in the genomes of RI strains, leading to the development of diabetic phenotypes. Recently, we examined and reported on the sensitivity of diabetes development to a high-fat diet in SMXA-5 mice and in the parental strains (SM/J and A/J) [20]. In that study, short-term feeding of a high-fat diet markedly worsened glucose tolerance in SMXA-5 mice. Thus, the SMXA-5 mouse is a diabetic model of susceptibility to a high-fat diet. On the other hand, it was demonstrated that the SM/J mouse is resistant to the induction of diabetes by a high-fat diet.
In this study, using the SMXA-5 and SM/J strains, which have different diabetes-related traits, we dissected diabetogenic loci in the A/J regions of the SMXA-5 genome that contribute to diabetes-related traits and obesity, such as impaired glucose tolerance, elevated blood glucose and insulin concentrations, and BMI. We attempted QTL analysis in mice of the (SM/J×SMXA-5)F2 intercross fed a high-fat diet, because such a diet enhances the differences in diabetes-related traits between SM/J and SMXA-5 mice. Moreover, to verify the function of the responsible locus that was mapped in the present study, we chose congenic mapping as a subsequent strategy. Congenic strains eliminate much of the genetic background interference and are efficient tools for dissecting complex diseases. Thus, we constructed a congenic strain by a marker-assisted method [21] and characterised the diabetes-related traits of this congenic strain.
Materials and methods
Parental strains, F1 hybrid and F2 intercross mice
Parental SM/J and SMXA-5 strains were obtained from the Institute for Laboratory Animal Research, Nagoya University School of Medicine (Nagoya, Japan). SM/J female mice were mated to SMXA-5 males to produce F1 hybrid mice, which were intercrossed to produce the (SM×SMXA-5)F2 intercross mice. Male F1 and F2 generations (a total of 255 mice) were produced and maintained in our facility. All mice were maintained in a room under conventional conditions at a controlled temperature of 23±3°C and 55±5% humidity on a 12-h light/12-h dark cycle. From 6 to 12 weeks, all mice were fed a high-fat diet. The mice were given free access to drinking water and the high-fat diet, the composition of which (g/kg diet) was as follows: casein, 209; carbohydrate (starch: sucrose, 1:1), 369; AIN93MX mineral mixture [22], 35; AIN93VX vitamin mixture [22], 10; choline chloride, 2; corn oil, 35; lard, 300; cellulose powder (Avicel type FD-101; Asahi Chemical Industry, Osaka, Japan), 40. The percentage of fat in this high-fat diet was 33.5 by weight. All procedures were performed in accordance with the Animal Experimental Guide of Nagoya University.
Production of congenic strain
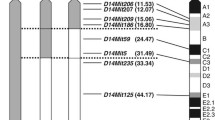
Male SM/J mice were mated to A/J female mice to produce F1 mice. Male F1 mice were backcrossed to SM/J females to produce the N2 generation. Male heterozygous carriers of chromosome 2 intervals between D2Mit295 and D2Mit281 were continuously bred to SM/J mice to produce the next generation. The marker-assisted ‘speed’ congenic procedure resulted in >99% replacement of the SM/J background genome by the N5 generation. The 154 microsatellite markers polymorphic between strains SM/J and A/J were used in the speed congenic procedure. N6 congenic animals were intercrossed to produce F1 animals (N6:F1) that were homozygous for the introgressed D2Mit295–D2Mit281 region from the A/J strain. The markers used in genotyping for the introgressed chromosomal region are shown in Fig. 1.
LOD score curves for blood glucose concentrations at 0 min (broken line) and 120 min (bold line) during IPGTT, as well as for the free-fed blood glucose concentration (thin line) and BMI (dotted/dashed line) for chromosome 2, and the chromosomal construction of SM.A-T2dm2sa congenic strain. Grey bar: introgressed chromosomal region from A/J into the SM.A-T2dm2sa congenic strain. Map positions (Mb) on the x-axis are taken from the Ensembl Genome Browser (http://www.ensembl.org/). Positions of four markers (D2Mit156, D2Mit162, D2Mit258 and D2Mit28) were not obtained. Suggestive, significant or highly significant thresholds for each trait were as follows: for IPGTT, fasting (1.9, 3.2, 4.7); 120 min (1.8, 3.2, 5.0); free-fed blood glucose (1.9, 3.3, 4.3); BMI (1.9, 3.3, 5.0)
Analytical procedures for phenotype determination
In this study, male mice were used in all experiments. In the experiment using F1 and F2 mice, all phenotype determinations were performed after 6–7 weeks of feeding on the high-fat diet, and mice were killed and dissected after 7 weeks. In the experiment using SM/J, A/J, SMXA-5 and SM.A-T2dm2sa mice, phenotype determinations were performed twice, after 6 and 10 weeks of feeding on the high-fat diet, and mice were killed and dissected after 11 weeks. BMI was calculated as body weight (g) divided by the square of the anal–nasal length (cm). Blood samples were obtained from the tail vein in free-fed or fasting mice at 09.00 h, and used for the measurement of blood glucose and insulin concentrations. Serum samples were collected, centrifuged and stored at −30°C until assay. Serum immunoreactive insulin concentrations were measured by radioimmunoassay (ShionoRIA; Shionogi, Osaka, Japan) with rat insulin as a standard. The intraperitoneal glucose tolerance test (IPGTT) was performed using the following protocol. After 14 h of fasting (from 19.00 to 09.00 h), blood samples were collected from the tail vein (fasting blood sample, 0 min sample in IPGTT). Then, a 20% glucose solution was injected intraperitoneally (2 g glucose/kg body weight). Blood samples were collected 30, 60 and 120 min after the injection. Blood glucose concentrations were measured by a glucose oxidase method (Glucose-B Test Kit; Wako, Osaka, Japan). The AUC was calculated according to the trapezoid rule from the glucose measurements at fasting, 30, 60 and 120 min (mmol min−1 l−1). Immediately after the mice in each experiment had been killed, the liver, subcutaneous fat and mesenteric fat were dissected and weighed. The anterior subcutaneous fat pad was dissected as subcutaneous fat tissue.
Genotyping and linkage analysis
Genomic DNA was prepared from mouse kidneys by salt–ethanol precipitation. A total of 73 microsatellite marker loci (Map Pair, Research Genetics, Huntsville, AL, USA), polymorphic between SM/J and SMXA-5, were genotyped in all 255 F2 mice. All microsatellite markers used and their locations obtained from the recombination frequencies in (SM/J×SMXA-5)F2 intercross mice are shown in Table S1 (Electronic Supplementary Material).
SMXA-5 is one of the 26 SMXA RI substrains that possess mosaic genomes derived from the SM/J and A/J strains [18]. We selected useful markers for this study from the SMXA RI strain distribution patterns [23]. PCRs were performed according to standard methods [24]. PCR products were separated by electrophoresis on a 4% NuSieve (FMC, Rockland, ME, USA) agarose gel and visualised by ethidium bromide staining. Linkage analysis was performed using the MapManager QTXb20 [25, 26] software program. This program is based on interval mapping using the free regression model. The permutation test estimates an empirical genome-wide probability for given likelihood ratio statistics (LRS). Significance was determined by 1-centimorgan (cM) steps for 1,000 permutations to provide LRS that were suggestive, significant or highly significant [27, 28]. The designations ‘suggestive’, ‘significant’ and ‘highly significant’ correspond to the 37th, 95th and 99.9th percentiles, respectively. The logarithm of the odds (LOD) score was obtained by dividing the LRS by 4.605 [29]. Significant linkage was defined in accordance with the guidelines of Lander and Kruglyak [30] as statistical evidence occurring by chance in the genome scan with p<0.05. The positions for all microsatellite markers were collected from the Ensembl Genome Browser (http://www.ensembl.org).
Statistical analysis
One-way ANOVA and subsequent Scheffé’s F test were used to compare the means among SM/J, SMXA-5 and F1 mice and among SM/J, A/J and SMXA-5 mice. Student’s t test was used to compare means between SM/J and SM.A-T2dm2sa mice. Differences for which the p value was <0.05 were regarded as significant in one-way ANOVA, Scheffé’s F test and Student’s t test. All of the statistical analyses were performed using StatView version 5.0 software (SAS Institute, Cary, NC, USA).
Results
Diabetes-related phenotypes in parental strains, F1, and F2 mice
In body weight, BMI and all of the diabetes-related phenotypes, the means in the SMXA-5 mice were significantly higher than those in the SM/J (Table 1). SMXA-5 showed mild obesity, markedly impaired glucose tolerance, hyperglycaemia and hyperinsulinaemia, whereas the SM/J mice showed none of these diabetic phenotypes (see Table 1 and a previous report [20]). The body weight and BMI of F1 mice were similar to those of SM/J mice. Blood glucose concentrations on fasting and at 30 and 60 min during the IPGTT and the AUC of F1 mice were significantly higher than those of SM/J mice. In contrast, blood glucose concentrations at fasting and at 60 and 120 min during the IPGTT and the AUC of F1 mice were significantly lower than those of SMXA-5 mice. The free-fed blood glucose concentration and the free-fed serum insulin concentration of F1 mice were similar to those of SM/J mice and lower than those of SMXA-5 mice. Most of these values in F1 mice lay between those of SM/J and SMXA-5 mice. In the F2 intercross mice, the means of all parameters, excluding blood glucose concentration at 30 min during the IPGTT, lay between those of SM/J and SMXA-5 mice (Table 1). Blood glucose concentrations at 120 min during the IPGTT of each of the SM/J, SMXA-5, F1 and F2 mice are plotted in Fig. 2. For all parameters, the individual values in F2 mice showed wide distributions, exceeding the ranges of values in SM/J and SMXA-5 mice (Table 1, Fig. 2).
QTL mapping in (SM×SMXA-5)F2 intercross
On chromosome 2, between D2Mit182 and D2Mit15, we mapped significant QTLs that affected all traits analysed in this study, except free-fed insulin concentration. A QTL for IPGTT at 120 min was found in the region near D2Mit15, the highest LOD score being 12.6 (significant LOD score=3.2). This locus was designated T2dm2sa, for type 2 diabetes mellitus 2 in the SMXA RI strains. T2dm2sa explains 9–20% of the phenotypic variance in blood glucose concentration at all time points during IPGTT (Table 2). The mean value of IPGTT at 120 min among F2 mice with the A/J homozygous genotype (A/A) at D2Mit15 was higher than the value in those with the SM/J homozygous genotype (SM/SM) (Table 2). On chromosome 2, the A/J allele at T2dm2sa increased the value of each phenotype based on the genotype in comparison with the SM/J allele. Each LOD score curve showed more than one peak for IPGTT at 120 min, fasting blood glucose concentration, free-fed blood glucose concentration, and BMI (Fig. 1). Concerning IPGTT at 120 min, there were large chromosomal regions exceeding significant thresholds of linkage.
On chromosome 12, we mapped significant and suggestive QTLs that affected BMI, IPGTT at 60 min, 120 min, AUC and free-fed blood glucose, fasting serum insulin, and free-fed serum insulin between D12Mit58 and D12Mit112 (Table 2). The mean values of these traits among F2 mice with the A/J homozygous genotype (A/A) between D12Mit58 and D12Mit112 were higher than those with the SM/J homozygous genotype (SM/SM) (Table 2). The diabetogenic allele originated in A/J was associated with increased value of each phenotype based on genotype.
In this study, significant and highly significant QTLs that affect diabetes-related traits and obesity were detected on only two chromosomes (chromosomes 2 and 12). However, we mapped suggestive QTLs for the diabetes-related traits and obesity on chromosomes 2, 5, 8, 11, 12, 14, 15, 17 and 19. We mapped suggestive QTLs on chromosome 5 for free-fed blood glucose; on chromosome 8 for fasting blood glucose; on chromosome 11 for BMI and glucose tolerance; on chromosome 14, for glucose tolerance; on chromosome 15 for fasting serum insulin; on chromosome 17 for BMI and glucose tolerance; and on chromosome 19 for glucose tolerance (Table 2). Each locus explains 3–6% of the phenotypic variance in obesity and diabetes-related traits. The diabetogenic allele that originated in A/J mice on chromosome 8, chromosome 14 and chromosome 19 was associated with increased values of each phenotype based on the genotype. In contrast, the diabetogenic allele originated in SM/J mice on chromosome 11 and 15 was associated with increased values of each phenotype based on the genotype. At the other QTLs on chromosome 17, the mean values of the traits in F2 mice with the heterozygous genotype were higher than the corresponding mean values in mice with the SM/J or A/J homozygous genotype.
Phenotype of congenic strain SM.A-T2dm2sa
The concept that the chromosomal region between D2Mit295 and D2Mit281, a segment of at least 120 Mb, contains the QTL for glucose tolerance is supported by significant thresholds of the LOD for IPGTT at 120 min (Fig. 1). The body weight and BMI of SM/J mice were significantly lower than those of both A/J and SMXA-5 mice. The body weight and BMI of SM.A-T2dm2sa mice were significantly higher than those of SM/J mice (Table 3). The blood glucose concentrations of SM/J mice during fasting and at 30 and 60 min during IPGTT were lower than those of A/J mice (p<0.0001, p=0.0393 and p=0.0096, respectively), but the values at 120 min were not significantly different (Fig. 3). The blood glucose concentrations of the SM.A-T2dm2sa mice during fasting and at 30, 60 and 120 min during IPGTT were significantly higher than those of SM/J. The impairment of glucose tolerance in SM.A-T2dm2sa mice appeared after 10 weeks on the high-fat diet, but was not observed after 6 weeks of feeding (data not shown). The free-fed blood glucose concentration of SM/J mice was lower than those of both A/J and SMXA-5 mice. The free-fed blood glucose concentration of SM.A-T2dm2sa mice was higher than that of SM/J mice (Table 3). The free-fed serum insulin concentration of SM/J mice was lower than that of SMXA-5 mice, but not lower than that of A/J mice (Table 3). The free-fed serum insulin concentration was higher in SM.A-T2dm2sa mice than in SM/J mice. The weight percentages of liver and subcutaneous fat of SM/J and A/J mice were significantly lower than the respective values of SMXA-5 mice. The weight percentage of mesenteric fat of SM/J mice was significantly lower than those of both A/J and SMXA-5 mice. The weight percentages of subcutaneous and mesenteric fat of SM.A-T2dm2sa mice were significantly higher than those of SM/J mice.
Glucose tolerance test in SM/J (squares), A/J (triangles), SMXA-5 (open circles) and SM.A-T2dm2sa (filled circles) congenic mice after 10 weeks of feeding. Data are mean±SEM. Means not sharing a common superscript letter are significantly different among SM/J, A/J and SMXA-5 by Scheffé’s F test (p<0.05). Means for SM/J and SM.A-T2dm2sa congenic mice were compared using Student’s t-test. * p<0.05 for difference from SM/J mice by Student’s t-test. p values of each time point during glucose tolerance test are as follows: fasting blood glucose, p<0.0001; 30 min, p=0.0002; 60 min, p<0.0001; 120 min, p<0.0001
Discussion
The common forms of human type 2 diabetes are caused by the interaction of multiple genes and environmental factors. In the present study we attempted the genetic dissection of loci contributing to impaired glucose tolerance, hyperglycaemia, hyperinsulinaemia and obesity by using the SMXA-5 mouse, which develops diabetes by the coexistence of diabetogenic alleles originating in the SM/J and A/J strains, under feeding on a high-fat diet. Although about half of the SMXA-5 genome is composed of the SM/J genome, the SMXA-5 mouse is susceptible to diabetes induced by a high-fat diet, whereas the SM/J mouse is resistant. Therefore, we decided to use the intercross between SMXA-5 and SM/J mice for QTL analysis. Using this intercross, the diabetogenic loci existing in the A/J regions of the SMXA-5 genome can be analysed, because the SMXA-5 has a mosaic genome derived from the non-diabetic SM/J and A/J strains.
QTL analysis for free-fed blood glucose concentration and glucose tolerance revealed the existence of a highly significant QTL on chromosome 2: T2dm2sa. We previously reported that QTL analysis of 19 SMXA RI strains fed a high-carbohydrate diet revealed several diabetogenic loci on four chromosomes (chromosomes 2, 6, 10 and 18), and the SMXA-5 mouse seems to possess these loci [19]. Except for the QTL on chromosome 2, the diabetogenic QTLs identified by using 19 SMXA RI strains were different from those identified in this study using (SM×SMXA-5)F2 mice. With regard to glucose tolerance, free-fed blood glucose concentration and serum insulin concentration, this study identified significant and suggestive QTLs on nine chromosomes. Moreover, the present results indicate that the QTL on chromosome 2, T2dm2sa, contributes the most strongly among these QTLs to the development of diabetes-related traits, but the effect of the QTL on chromosome 2 on these traits was weak among 19 SMXA RI strains. A high-fat diet might strengthen the effect of T2dm2sa on the development of diabetes-related traits in (SM×SMXA-5)F2 mice.
Using different polygenic type 2 diabetes mouse models, three groups have reported diabetes-related QTLs on chromosome 2 near T2dm2sa, which we mapped in this study (D2Mit15; 50 cM, 92.7 Mb). Firstly, Hirayama et al. reported Nidd5, a QTL that affects body weight and plasma insulin concentration, by using (TSOD×BALB/c)F2 intercross mice [12]. Secondly, Mehrabian et al. reported Mob (multigenic obesity) loci, a QTL that affects body weight, subcutaneous fat mass and plasma insulin, by using (C57BL/6×CAST/Ei)F2 intercross mice [31]. In addition, three congenic mouse strains with Mob loci were created, and differences in obesity and insulin traits consistent with the original QTL analysis were shown [32]. Thirdly, Stoehr et al. identified T2dm3, a QTL that affects the fasting plasma insulin concentration, by using a (BTBR-+/+×C57BL/6J-ob/+)F2 intercross [33], and mapped Moo1 (modifier of obese) in the centromeric region of T2dm3, a QTL that controls body mass [34]. T2dm2sa also has suggestive or significant linkage to free-fed insulin concentration or fasting insulin concentration, respectively, as did Nidd5, Mob and T2dm3. Although these three QTLs and T2dm2sa were mapped on the middle of mouse chromosome 2 by using different diabetic mouse strains, it is unclear at present whether T2dm2sa possesses the same diabetogenic gene that these QTLs do.
The chromosomal region with significant linkage of blood glucose concentration at 120 min during IPGTT is homologous to human chromosomes 2q11–q32, 9q33–34, 11p14–q12, 15q13–q21 and 20p13–11, and to rat chromosome 3. Human chromosomes 2 [35] and 20 [36–39] encompass the region associated with insulin resistance. In the WOKW rat, the homologous region on rat chromosome 3 contains a QTL that affects AUC during IPGTT [40]. These mapping data in the human, rat, and mouse indicate that this homologous region may possess orthologous diabetogenic genes.
Nr1h3, a possible candidate gene in T2dm2sa, encodes liver X receptor-α (LXR-α). This transcription factor is known to play an important role in the regulation of cholesterol metabolism [41, 42]. Nr1h3 is expressed in liver, kidney, intestine, adipose tissue and adrenals. Recently, LXR-α was also reported to play an important role in the regulation of gluconeogenesis. In the db/db mouse and the insulin-resistant Zucker fa/fa rat, the activation of LXR by a specific agonist lowered the plasma glucose level [43] by suppressing the expression of genes involved in hepatic gluconeogenesis, including phosphoenolpyruvate carboxykinase (PEPCK). The gene expression profile in the liver of LXR-α and -β knockout mice showed that LXRs suppressed the expression of gluconeogenic genes encoding such substances as PEPCK, fructose-1,6-bisphosphatase, and glucose-6-phosphatase [43, 44]. In addition, LXR-α also regulates glucose uptake in peripheral tissue (muscle and adipose tissue) by upregulating the expression of the gene encoding GLUT4 [45, 46]. Thus, at present, we consider Nr1h3 the most plausible candidate gene for T2dm2sa in SMXA-5 mice.
On chromosome 12, we mapped a significant QTL that affects BMI and free-fed serum insulin but not glucose tolerance or blood glucose concentration. Therefore, this locus might be linked to obesity and insulin resistance. Two QTLs, Afpq10 (abdominal fat percent QTL 10; 18.0 cM) and Afw1 (abdominal fat weight QTL 10; 21.0 cM) were mapped around our QTL region by using crosses between DU6i mice and DBA/2 mice [47]. It is suggested that the gene commonly contributing to obesity may exist in the centromeric region of chromosome 12. However, there is no QTL for obesity on chromosome 12 in the studies using SMXA RI substrains or (SM×A)F2 mice [19, 48, 49]. Therefore, this QTL may be a locus specifically activated under a high-fat diet.
For our next step in QTL analysis, we chose congenic mapping as a strategy to dissect T2dm2sa. The glucose tolerance, free-fed blood glucose concentration, BMI and accumulation of mesenteric fat of SM.A-T2dm2sa mice differed significantly from those of mice with the SM/J background. This result clearly verified that T2dm2sa exerts definitive effects on the development of diabetes-related phenotypes and obesity. Although A/J mice possess a diabetogenic allele at T2dm2sa, they show no obviously diabetic phenotypes, unlike SM.A-T2dm2sa mice. These findings suggest that the coexistence of the T2dm2sa and other diabetogenic loci, which exist outside T2dm2sa, leads to diabetes in SM.A-T2dm2sa mice. In the present QTL analysis in (SM×SMXA-5)F2 mice, diabetogenic loci within the SM/J regions of the SMXA-5 genome cannot be detected. Therefore, unknown diabetogenic loci in SMXA-5 mice may exist within the SM/J regions of the SMXA-5 genome. At present, we are trying to dissect such unknown diabetogenic loci involved in the development of diabetes in both SMXA-5 and SM.A-T2dm2sa mice.
In conclusion, we detected nine loci associated with several diabetes-related traits and/or obesity on chromosomes 2, 5, 8, 11, 12, 14, 15, 17 and 19 by QTL analysis in F2 intercross mice derived from SM/J and diabetic SMXA-5 mice. Locus T2dm2sa, which strongly affected glucose tolerance, blood glucose concentration and obesity, was mapped on chromosome 2. Moreover, although A/J (donor) and SM/J (recipient) are non-diabetic strains, the congenic strain, SM.A-T2dm2sa, developed overt impaired glucose tolerance. The results of the present study suggest that both SM/J and A/J mouse strains possess latent diabetic genes and that the coexistence of these alleles is necessary for eliciting diabetes in SMXA-5 mice. The dissection of diabetogenic genes of this kind, which have epistatic effects, will contribute to the elucidation of the complex mechanisms underlying human diabetes.
Abbreviations
- cM:
-
centimorgan
- IPGTT:
-
intraperitoneal glucose tolerance test
- LOD:
-
logarithm of the odds
- LXR-α:
-
liver X receptor-α
- PEPCK:
-
phosphoenolpyruvate carboxykinase
- QTL:
-
quantitative trait locus
- RI:
-
recombinant inbred
- T2dm2sa :
-
gene for type 2 diabetes mellitus 2 in SMXA RI mice
References
Warm JH, Rich SS, Krolewski AS (1994) Epidemiology and genetics of diabetes mellitus. In: Kahn C, Weir G (eds) Joslin’s diabetes mellitus. Lea and Febiger, Philadelphia, Pennsylvania, pp 201–215
Galli J, Li LS, Glaser A et al (1996) Genetic analysis of non-insulin dependent diabetes mellitus in the GK rat. Nat Genet 12:31–37
Gauguier D, Froguel P, Parent V et al (1996) Chromosomal mapping of genetic loci associated with non-insulin dependent diabetes in the GK rat. Nat Genet 12:38–43
Kanemoto N, Hishigaki H, Miyakita A et al (1998) Genetic dissection of ‘OLETF’, a rat model for non-insulin-dependent diabetes mellitus. Mamm Genome 9:419–425
Moralejo DH, Ogino T, Zhu M et al (1998) A major quantitative trait locus co-localizing with cholecystokinin type A receptor gene influences poor pancreatic proliferation in a spontaneously diabetogenic rat. Mamm Genome 9:794–798
Wei S, Wei K, Moralejo DH et al (1999) Mapping and characterization of quantitative trait loci for non-insulin-dependent diabetes mellitus with an improved genetic map in the Otsuka Long-Evans Tokushima fatty rat. Mamm Genome 10:249–258
Sugiura K, Miyake T, Taniguchi Y et al (1999) Identification of novel non-insulin-dependent diabetes mellitus susceptibility loci in the Otsuka Long-Evans Tokushima fatty rat by MQM-mapping method. Mamm Genome 10:1126–1131
Masuyama T, Fuse M, Yokoi N et al (2003) Genetic analysis for diabetes in a new rat model of nonobese type 2 diabetes, spontaneously diabetic torii rat. Biochem Biophys Res Commun 304:196–206
Ueda H, Ikegami H, Kawaguchi Y et al (1999) Genetic analysis of late-onset type 2 diabetes in a mouse model of human complex trait. Diabetes 48:1168–1174
Leiter EH, Reifsnyder PC, Flurkey K, Partke HJ, Junger E, Herberg L (1998) NIDDM genes in mice: deleterious synergism by both parental genomes contributes to diabetogenic thresholds. Diabetes 47:1287–1295
Plum L, Giesen K, Kluge R et al (2002) Characterisation of the mouse diabetes susceptibility locus Nidd/SJL: islet cell destruction, interaction with the obesity QTL Nob1, and effect of dietary fat. Diabetologia 45:823–830
Hirayama I, Yi Z, Izumi S et al (1999) Genetic analysis of obese diabetes in the TSOD mouse. Diabetes 48:1183–1191
Shike T, Hirose S, Kobayashi M, Funabiki K, Shirai T, Tomino Y (2001) Susceptibility and negative epistatic loci contributing to type 2 diabetes and related phenotypes in a KK/Ta mouse model. Diabetes 50:1943–1948
Suto J, Matsuura S, Imamura K, Yamanaka H, Sekikawa K (1998) Genetics of obesity in KK mouse and effects of Ay allele on quantitative regulation. Mamm Genome 9:506–510
Suto J, Matsuura S, Imamura K, Yamanaka H, Sekikawa K (1998) Genetic analysis of non-insulin dependent diabetes mellitus in KK and KK-Ay mice. Eur J Endocrinol 139:654–661
Kim JH, Sen S, Avery CS et al (2001) Genetic analysis of a new mouse model for non-insulin-dependent diabetes. Genomics 74:273–286
Nishimura M, Hirayama N, Serikawa T et al (1995) The SMXA: a new set of recombinant inbred strain of mice consisting of 26 substrains and their genetic profile. Mamm Genome 6:850–857
Anunciado RV, Horio F, Ohno T, Tanaka S, Nishimura M, Namikawa T (2000) Characterization of hyperinsulinemic recombinant inbred (RI) strains (SMXA-5 and SMXA-9) derived from normoinsulinemic SM/J and A/J mice. Exp Anim 49:83–90
Kobayashi M, Ohno T, Tsuji A, Nishimura M, Horio F (2003) Combinations of nondiabetic parental genomes elicit impaired glucose tolerance in mouse SMXA recombinant inbred strains. Diabetes 52:180–186
Kobayashi M, Io F, Kawai T, Nishimura M, Ohno T, Horio F (2004) SMXA-5 mouse as a diabetic model susceptible to feeding a high-fat diet. Biosci Biotechnol Biochem 68:226–230
Markel P, Shu P, Ebeling C et al (1997) Theoretical and empirical issues for marker-assisted breeding of congenic mouse strains. Nat Genet 17:280–284
Reeves PG, Nielsen FH, Fahey GC Jr (1993) AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J Nutr 123:1939–1951
Ohno T, Katoh J, Kikkawa Y, Yonekawa H, Nishimura M (2003) Improved strain distribution patterns of SMXA recombinant inbred strains by microsatellite markers. Exp Anim 52:415–417
Silver LM (1995) Genetic markers. In: Silver LM (ed.) Mouse genetics. Oxford University Press, New York, pp 184–190
Manly KF, Olson JM (1999) Overview of QTL mapping software and introduction to map manager QT. Mamm Genome 10:327–334
Manly KF, Cudmore RH Jr, Meer JM (2001) Map Manager QTX, cross-platform software for genetic mapping. Mamm Genome 12:930–932
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Doerge RW, Churchill GA (1996) Permutation tests for multiple loci affecting a quantitative character. Genetics 142:285–294
Lynch M, Walsh B (1997) Mapping and characterizing QTLs: inbred line crosses. In: Genetics and analysis of quantitative traits. Sinauer Associates, Sunderland, Massachusetts, pp 431–489
Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247
Mehrabian M, Wen PZ, Fisler J, Davis RC, Lusis AJ (1998) Genetic loci controlling body fat, lipoprotein metabolism, and insulin levels in a multifactorial mouse model. J Clin Invest 101:2485–2496
Estrada-Smith D, Castellani LW, Wong H et al (2004) Dissection of multigenic obesity traits in congenic mouse strains. Mamm Genome 15:14–22
Stoehr JP, Nadler ST, Schueler KI et al (2000) Genetic obesity unmasks nonlinear interactions between murine type 2 diabetes susceptibility loci. Diabetes 49:1946–1954
Stoehr JP, Byers JE, Clee SM et al (2004) Identification of major quantitative trait loci controlling body weight variation in ob/ob mice. Diabetes 53:245–249
Young TL, Penny L, Woods MO et al (1999) A fifth locus for Bardet–Biedl syndrome maps to chromosome 2q31. Am J Hum Genet 64:900–904
Beales PL, Katsanis N, Lewis RA et al (2001) Genetic and mutational analyses of a large multiethnic Bardet–Biedl cohort reveal a minor involvement of BBS6 and delineate the critical intervals of other loci. Am J Hum Genet 68:606–616
Zouali H, Hani EH, Philippi A et al (1997) A susceptibility locus for early-onset non-insulin dependent (type 2) diabetes mellitus maps to chromosome 20q, proximal to the phosphoenolpyruvate carboxykinase gene. Hum Mol Genet 6:1401–1408
Hunt SC, Abkevich V, Hensel CH et al (2001) Linkage of body mass index to chromosome 20 in Utah pedigrees. Hum Genet 109:279–285
Permutt MA, Wasson J, Love-Gregory L et al (2002) Searching for type 2 diabetes genes on chromosome 20. Diabetes 51 (Suppl 3):S308–S315
Kovács P, van den Brandt J, Klöting I (2000) Genetic dissection of the syndrome X in the rat. Biochem Biophys Res Commun 269:660–665
Lehmann JM, Kliewer SA, Moore LB et al (1997) Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway. J Biol Chem 272:3137–3140
Peet DJ, Turley SD, Ma W et al (1998) Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell 93:693–704
Cao G, Liang Y, Broderick CL et al (2003) Antidiabetic action of a liver X receptor agonist mediated by inhibition of hepatic gluconeogenesis. J Biol Chem 278:1131–1136
Stulnig TM, Steffensen KR, Gao H et al (2002) Novel roles of liver X receptors exposed by gene expression profiling in liver and adipose tissue. Mol Pharmacol 62:1299–1305
Laffitte BA, Chao LC, Li J et al (2003) Activation of liver X receptor improves glucose tolerance through coordinate regulation of glucose metabolism in liver and adipose tissue. Proc Natl Acad Sci USA 100:5419–5424
Dalen KT, Ulven SM, Bamberg K, Gustafsson JA, Nebb HI (2003) Expression of the insulin-responsive glucose transporter GLUT4 in adipocytes is dependent on liver X receptor alpha. J Biol Chem 278:48283–48291
Brockmann GA, Kratzsch J, Haley CS, Renne U, Schwerin M, Karle S (2000) Single QTL effects, epistasis, and pleiotropy account for two-thirds of the phenotypic F2 variance of growth and obesity in DU6i × DBA/2 mice. Genome Res 10:1941–1957
Anunciado RV, Ohno T, Mori M et al (2000) Distribution of body weight, blood insulin and lipid levels in the SMXA recombinant inbred strains and the QTL analysis. Exp Anim 49:217–224
Anunciado RV, Nishimura M, Mori M et al (2003) Quantitative trait locus analysis of serum insulin, triglyceride, total cholesterol and phospholipid levels in the (SM/J × A/J)F2 mice. Exp Anim 52:37–42
Acknowledgements
This work was supported by a Grant-in-Aid for Scientific Research (C) (No. 15580105) from the Japan Society for the Promotion of Science, a grant from Kao Research Council for the Study of Healthcare Science, Japan, and by a grant from the Elizabeth Arnold Fuji Foundation, Japan. We thank T. Toyoda, N. Heida and Y. Hasegawa of the Genomic Knowledge Base Research Team, Bioinformatics Group, Genomic Sciences Center, RIKEN, for invaluable discussion on bioinformatics. We thank A. Ishikawa of the Laboratory of Animal Genetics, Nagoya University for his helpful comments on genetic analysis. We thank T. Kawada of the Division of Food Science and Biotechnology, Kyoto University, for his helpful comments on diabetes-related traits and obesity. M. Kobayashi is a Research Fellow of the Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary materials
Rights and permissions
About this article
Cite this article
Kobayashi, M., Io, F., Kawai, T. et al. Major quantitative trait locus on chromosome 2 for glucose tolerance in diabetic SMXA-5 mouse established from non-diabetic SM/J and A/J strains. Diabetologia 49, 486–495 (2006). https://doi.org/10.1007/s00125-005-0121-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-005-0121-3