Abstract
Aims/hypothesis
Resistin is a peptide secreted by adipocytes and recognized as a hormone that could link obesity to insulin resistance. This study was designed to examine the effect and mechanism(s) of insulin on resistin expression in 3T3-L1 adipocytes.
Methods
Differentiated 3T3-L1 adipocytes were stimulated with insulin and resistin mRNA expression was examined by Northern blot analysis. In some experiments, the insulin signal was blocked by several chemical inhibitors or overexpression of a dominant negative form (Δp85) of the p85 subunit of phosphatidylinositol 3-kinase (PI 3-kinase).
Results
Insulin treatment caused a reduction of resistin mRNA in time-dependent and dose-dependent manners in 3T3-L1 adipocytes. Pre-treatment with PD98059, an inhibitor of extracellular signal-regulated kinase 1/2 (ERK1/2) pathway, or SB203580, an inhibitor of p38 mitogen-activated protein-kinase (p38 MAP-kinase) pathway, did not influence insulin-induced reduction of resistin mRNA. Inhibition of PI 3-kinase by LY294002 or Δp85 also failed to block insulin-induced reduction of resistin mRNA. Cycloheximide, a protein synthesis inhibitor, completely blocked insulin-induced reduction of resistin mRNA. Actinomycin D, a RNA synthesis inhibitor, also blocked insulin-induced reduction of resistin mRNA, and the decreasing rate of resistin mRNA in cells treated with insulin alone was faster than that with actinomycin D.
Conclusion/interpretation
Insulin downregulates resistin mRNA via PI 3-kinase, ERK or p38 MAP-kinase independent pathways in 3T3-L1 adipocytes. The downregulation mechanism of resistin mRNA by insulin would be an indirect event through the synthesis of novel protein(s) that could accelerate the degradation of resistin mRNA.
Similar content being viewed by others
Type 2 diabetes mellitus is a polygenic disorder in which insulin resistance in peripheral tissues and the failure of insulin secretion from pancreatic beta cells are two major aetiologic factors [1]. Insulin resistance is often associated with obesity, however, the mechanism by which obesity results in the impairment of insulin sensitivity is not completely understood [2]. It is known that increase of serum free fatty acids (FFA), lipids produced from adipose tissue, could worsen insulin sensitivity by impairing insulin-signalling molecules [2]. It has been shown that the adipose tissue secretes several hormones and cytokines, and that some of these factors, such as tumour necrosis factor-α (TNF-α), leptin and adiponectin, influence insulin resistance both in vivo and in vitro [3, 4, 5].
Resistin, also known as an adipose tissue-specific secretory factor (ADSF), is a novel cysteine-rich protein secreted from differentiated 3T3-L1 adipocytes and white adipose tissue [6, 7]. Previous studies have shown that administration of resistin protein in mice worsens insulin sensitivity, whereas treatment with anti-resistin antibody partially restores insulin sensitivity in obese mice, suggesting that resistin is a potential factor that mediates insulin resistance [6]. It has been reported that serum resistin concentrations are increased in mice with diet-induced obesity as well as in genetic models of obesity, ob/ob and db/db mice [6]. In contrast, another study [8] has recently reported the opposite result, i.e., resistin mRNA is decreased in obese animals including the ob/ob, db/db, tub/tub, and KKAy mice compared with the respective lean counterparts.
In man, clinical studies have shown that resistin mRNA expression in adipose tissues are increased in obese humans [9] and that SNPs in the resistin gene promoter are associated with obesity [10]. However, there are controversial reports that show the lack of correlation between obesity and resistin expression in adipocytes [9, 11, 12], and therefore it is not clear whether resistin affects insulin resistance in humans.
Little is known about the control of resistin gene expression except in those reports that show inhibition of resistin gene expression by isoproterenol, a β-adrenergic receptor agonist [13] and TNF-α in 3T3-L1 adipocytes [14]. It is well known that insulin is an important regulator of the expression of genes involved in metabolism like those of phosphoenolpyruvate carboxykinase (PEPCK) and fatty acid synthase [15]. Another study. [16] has shown that insulin could reduce the resistin mRNA expression in 3T3-L1 adipocytes. This is surprising since serum resistin concentrations in mice decrease after 48-h fasting, and reverse to normal concentrations after feeding, with a parallel change in the expression of resistin mRNA in adipose tissues [6] and insulin treatment of diabetic animals causes an increase in resistin expression [7]. In this study, we used differentiated and undifferentiated 3T3-L1 cells, and investigated the impact of insulin on the regulation of resistin mRNA expression and potential insulin signalling pathways that are involved in this control process.
Materials and methods
Cell culture
3T3-L1 fibroblasts were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% calf serum (Invitrogen, Carlsbad, Calif., USA) and 25 mmol/l glucose in an atmosphere of 10% CO2 at 37°C. 3T3-L1 fibroblasts differentiated into adipocytes after reaching confluence by the addition of differentiation medium [DMEM containing 10% foetal bovine serum (FBS), 25 mmol/l glucose, 0.5 mmol/l 3-isobutyl-1-methylxanthine, 5 µg/ml insulin and 0.4 µg/ml dexamethasone]. After 48 h, the culture medium was replaced with adipocyte growth medium (DMEM containing 10% FBS, 25 mmol/l glucose and 5 µg/ml insulin) and maintained for another 48 h. The cells were then cultured in DMEM supplemented with 10% FBS and 25 mmol/l glucose for another 4 to 12 days.
Northern blot analysis
For all experiments, differentiated adipocytes at day 8 were serum deprived for 6 h in DMEM medium containing 25 mmol/l glucose and 0.1% bovine serum albumin (BSA) and then, unless noted otherwise, incubated with or without insulin at a concentration of 100 nmol/l in DMEM medium supplemented with 25 mmol/l glucose and 0.1% BSA for the indicated time periods. In some cases, cells were preincubated with 50 µmol/l of PD98059 (Calbiochem, San Diego, Calif., USA), 10 µmol/l of SB203580 (Calbiochem) or 50 µmol/l LY294002 (Sigma, St. Louis, Mo., USA) for 30 min and stimulated with insulin for 16 h with each inhibitor. Cells were preincubated with 5 µg/ml of cycloheximide (Sigma) or 5 µg/ml of actinomycin D (Sigma) for 30 min and stimulated with insulin for 16 h with each inhibitor. Total RNA was isolated using TRIzol reagent (Invitrogen). Northern blot analysis was carried out according to standard techniques in denaturing formaldehyde-containing agarose gels. Total RNA (10 µg) was subjected to electrophoresis on 1.0% agarose gels. Ethidium bromide staining of the gels confirmed equal loading and integrity of the RNA. After transfer to Hybond-N+ (Amersham Pharmacia Biotech, UK), blots were hybridised in ExpressHyb Hybridization Solution (Clontech Laboratories, Palo Alto, Calif., USA) with [α-32P] dCTP-labelled probes corresponding to mouse resistin cDNA (from up 138 to 421 [6]), mouse aP2 cDNA (from up 1 to 399 [17]) or mouse 36B4 cDNA (from up 336 to 628 [18]). After incubation for 2 h at 68°C, blots were washed twice in 2×SSC buffer (300 mmol/l NaCl and 30 mmol/l sodium citrate) with 0.05% SDS for 10 min each at room temperature and for 10 min in 0.1×SSC with 0.1% SDS at 50°C. Membranes were then subjected to autoradiography.
Western blot analysis
Cells were pretreated with PD98059 or LY294002 as described above in Northern Blot analysis. They were then stimulated with or without 100 nmol/l insulin for 10 min with each inhibitor. Whole cell protein extracts were prepared by using buffer A [30 mmol/l Tris (pH 7.4), 150 mmol/l NaCl, 10 mmol/l EDTA, 1 mmol/l Na3VO4, 20 mmol/l Na4P2O2, 50 mmol/l NaF, 1% NP-40, 1 mmol/l phenylmethylsulfonyl fluoride (PMSF), 10 µg/ml of aprotinin, and 1 µmol/l leupeptin] for 30 min at 4°C, and insoluble protein was removed by centrifugation at 12,000 rpm in a microcentrifuge. Protein content was calculated by the method of Bradford [19]. The extract was then resolved in SDS-polyacrylamide gels after boiling in Laemmli SDS sample buffer. Approximately 50 µg of protein samples were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose transfer membranes (Schleicher & Schuell, Keene, N.H., USA). The blots were blocked with 3% of BSA in TBS buffer [10 mmol/l Tris (pH 7.5) and 150 mmol/l NaCl] and then incubated with anti-ERK1/2 antibody (Santa Cruz Biotechnology, Santa Cruz, Calif., USA), anti-phospho-ERK1/2 antibody (Cell Signalling, Beverly, Mass., USA), anti-Akt antibody (Cell Signalling) or anti-phospho-Akt antibody (Cell Signalling) in TBS containing 2% BSA, followed by incubation with corresponding secondary antibodies conjugated to horseradish peroxidase. The immunoreactive bands were visualized by enhanced chemiluminescence quantified by densitometric analysis.
Assay of p38 MAP-kinase activity
Cells were pretreated with SB203580 for 30 min and stimulated with or without 100 nmol/l insulin for 10 min with SB203580. The activity of p38 MAP-kinase was measured using p38 MAP Kinase Assay Kit (Cell Signalling). Briefly, the cells were washed once with ice-cold phosphate-buffered saline and lysed in lysis buffer [20 mmol/l Tris (pH 7.4), 150 mmol/l NaCl, 1 mmol/l EDTA, 1 mmol/l EGTA, 1% Triton, 2.5 mmol/l sodium pyrophosphate, 1 mmol/l β-glycerolphosphate, 1 mmol/l Na3VO4, 1 µg/ml leupeptin]. The lysate was sonicated and centrifuged, and the supernatant was incubated with a re-suspended immobilised phospho-p38 MAP-kinase monoclonal antibody with gentle rocking for 12 h at 4°C. The beads were washed twice with 500 µl of lysis buffer and twice with 500 µl of kinase buffer [25 mmol/l Tris (pH 7.5), 5 mmol/l β-glycerol-phosphate, 2 mmol/l DTT, 0.1 mmol/l Na3VO4, and 10 mmol/l MgCl2]. The kinase reactions were carried out with 200 µmol/l ATP and 2 µg of ATF-2 at 30°C for 30 min. ATF-2 phosphorylation was selectively measured by Western blots using a chemiluminescent detection system and specific antibodies against phosphorylation of ATF-2 at Thr71.
Adenovirus-mediated gene transfer
Adenovirus vector encoding Δp85 that lack the binding site for the 110-kDa catalytic subunit of PI 3-kinase (AdexCAΔp85) was kindly provided by Dr. M. Kasuga (Kobe University, Kobe, Japan) [20]. Adenovirus vector encoding LacZ gene (AdexCALacZ) was kindly provided by Dr. K. Ueki (Joslin Diabetes Center, Boston, Mass., USA). 3T3-L1 adipocytes were infected with AdexCALacZ or AdexCAΔp85 in DMEM containing 10% FBS and 25 mmol/l glucose for 24 h. For Northern blot analysis, cells were serum starved for 6 h and then stimulated with insulin at a concentration of 100 nmol/l for 16 h. For PI 3-kinase assay, cells were serum starved for 6 h and then stimulated with insulin at a concentration of 100 nmol/l for 10 min.
PI 3-kinase assay
Cells were infected with AdexCALacZ or AdexCAΔp85 and, after 24 h, cells were serum starved for 6 h and stimulated with or without 100 nmol/l insulin for 10 min. Whole cell protein extracts were prepared as described above under Western blot analysis. Approximately 500 µg of protein samples were subjected to immunoprecipitation for 12 h using anti-phosphotyrosine antibody (Upstate, Lake Placid, N.Y., USA). Subsequently, protein A-Sepharose was added and followed by incubation for another 90 min. Immune-complexed PI 3-kinase activity was measured by phosphorylation of phosphatidylinositides (PI). Immune-complexes were incubated in 45 µl reaction buffer containing 10 µmol/l ATP, 1.1 MBq of [γ-32P] ATP and 10 µg of PI at room temperature under constant shaking. After 10 min, the reaction was stopped by the addition of 250 µl of 1 N HCl and 80 µl of CHCl3/methanol (2:1). The samples were centrifuged, and the lower organic phase was removed and applied to a silica gel TLC plate (Merck, Germany) coated with 1% potassium oxalate. TLC plates were developed in CHCl3/CH3OH/H2O/NH4OH (60:47:11.3:2), dried, and visualized by autoradiography. The 32P-labelled PI was quantified using NIH image analysis software.
Statistical analysis
Data are expressed as means ± standard error of the mean (SEM). Differences between two groups were evaluated by unpaired Student's t test. A p value of less than 0.05 was considered statistically significant.
Results
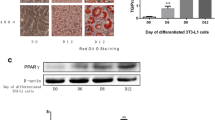
Firstly, we investigated resistin mRNA expression in 3T3-L1 cells during adipocyte differentiation. Resistin mRNA expression in pre-adipocytes (differentiation at days 0 and 2) was below the detection limit, but was detected from day 4. Resistin mRNA expression reached a maximum at day 8, but then decreased at day 12 (Fig. 1). aP2 mRNA, a marker of adipocyte differentiation, was weakly detected at day 2, reached a maximum at day 8, and remained at that expression until day 12.
Resistin mRNA expression in 3T3-L1 cells during adipocyte differentiation. 3T3-L1 fibroblasts were differentiated to adipocytes. Equal amount of total RNA (10 µg/lane) prepared from cells at the indicated time point was subjected to Northern blot analysis for resistin, aP2, and 36B4 mRNA (an internal control)
To examine the effect of insulin on resistin mRNA expression in differentiated adipocytes, 3T3-L1 adipocytes at 8 days after induction of differentiation were incubated with 100 nmol/l insulin for various time intervals (Fig. 2A,B). Insulin significantly decreased resistin mRNA expression at 8 h (p <0.05) and resulted in a further decrease to about 30% after 24 h, compared with that in non-insulin-treated cells (p <0.01). In contrast, the expression of aP2 mRNA was not altered by insulin (Fig. 2A). The effect of insulin on resistin mRNA was dose-dependent with a significant inhibition at concentrations as low as 10 nmol/l (p<0.05), but reached a plateau at 100 nmol/l insulin (Fig. 2C,D). At 100 nmol/l, insulin reduced resistin mRNA to 33% of that in non-treated cells (p<0.05).
Effect of insulin treatment on resistin mRNA expression in 3T3-L1 adipocytes. (A) After 6-h serum starvation, differentiated 3T3-L1 adipocytes (day 8) were incubated with insulin (100 nmol/l) for indicated time intervals. Total RNA was extracted from the cells and equal amount of RNA (10 µg/lane) was subjected to Northern blot analysis of resistin, aP2, and 36B4 mRNAs. (B)Quantification of the results. Resistin mRNA was corrected with 36B4 mRNA. Data, expressed as percent of resistin mRNA without insulin treatment (control), are expressed as the means ± SEM of three independent experiments. *p <0.05 vs. control. **p <0.01 vs. control. (C) Serum-starved 3T3-L1 adipocytes were incubated with insulin at indicated concentrations for 16 h. Total RNA samples (10 µg/lane) were examined by Northern blot analysis for resistin and 36B4 mRNA. (D) Quantification of the results. Resistin mRNA was corrected with 36B4 mRNA and expressed as mentioned above
We tested whether the insulin-induced reduction of resistin mRNA was modified by insulin-induced glucose uptake. For this purpose, cells were cultured with or without glucose (25 mmol/l) for 6 h and stimulated with insulin for 16 h. Without insulin stimulation but under a 25 mmol/l glucose condition, resistin mRNA expression was four times higher than that under glucose-free condition (Fig. 3A,B), indicating that extracellular glucose is another stimulus for resistin mRNA expression. However, insulin-induced decrease of resistin mRNA still occurred under glucose-free conditions (Fig. 3A,B), suggesting that glucose uptake through insulin stimulation is not essential for insulin-induced decrease of resistin mRNA.
Expression of resistin mRNA in cells cultured with glucose-free medium. (A) 3T3-L1 adipocytes were serum-starved for 6 h in DMEM medium containing 0.1% BSA with or without 25 mmol/l glucose and were stimulated with or without insulin for 16 h. Resistin mRNA was examined by Northern blot analysis. Equal load of the total RNA was confirmed by 28S/18S ribosomal RNA. (B) Resistin mRNA was expressed as a percent of resistin mRNA in cells cultured in 25 mmol/l glucose without insulin stimulation (control). Data are shown as the mean ± SEM of three independent experiments. *p <0.05 vs. control. †p <0.05 0 mmol/l glucose without insulin vs. 0 mmol/l glucose with insulin
Analyses of insulin signalling pathways
We then examined the downstream signals that could be involved in insulin-induced down-regulation of resistin mRNA. It is well known that regulation of gene transcription by insulin is mediated mainly via PI 3-kinase or ERK1/2, a member of MAP-kinase family. Firstly, we assessed the role of ERK1/2 in insulin-induced reduction of resistin mRNA by pre-treatment of the cells with PD98059, a chemical inhibitor of ERK1/2. 3T3-L1 adipocytes were pretreated with PD98059 (50 µmol/l) for 30 min, and then incubated with or without insulin (100 nmol/l) for 16 h with PD98059. PD98059 did not influence insulin-induced reduction of resistin mRNA (Fig. 4A), although ERK1/2 phosphorylation by insulin could be blocked by 50 µmol/l PD98059 (Fig. 4B).
Effects of PD98059 and SB203580 on insulin-induced decrease of resistin mRNA. (A, C) 3T3-L1 adipocytes were pretreated with or without PD98059 (A) or SB203580 (C) for 30 min and stimulated with insulin for 16 h. Resistin mRNA expression was examined by Northern blot analysis. Resistin mRNA was corrected with 36B4 mRNA. Data, expressed as percent of resistin mRNA in cells not treated with insulin or the inhibitor (control), are expressed as the means ± SEM of three independent experiments. *p<0.05 vs. control. †p<0.05 vs. each treatment. n.s.: no significant change. (B) 3T3-L1 adipocytes were pretreated with or without PD98059 for 30 min and stimulated with insulin for 10 min. Total cell lysates were subjected to Western blot analysis using anti-phospho-ERK1/2 antibody or anti-ERK1/2 antibody. (D) 3T3-L1 adipocytes were pretreated with or without SB203580 for 30 min and stimulated with insulin for 10 min. Cell lysates were subjected to p38 MAP-kinase activity assay. Data, expressed as percent of p38 MAP-kinase activity in cells not treated with insulin or SB203580 (control), are expressed as the means ± SEM of three independent experiments
It is also known that p38 MAP-kinase is phosphorylated and activated by insulin in 3T3-L1 adipocytes. To assess whether the p38 MAP-kinase pathway is involved in insulin-induced reduction of resistin mRNA, 3T3-L1 adipocytes were pretreated with 10 µmol/l SB203580, an inhibitor of p38 MAP-kinase pathway, for 30 min and then incubated with or without 100 nmol/l insulin for 16 h with SB203580. SB203580 did not block insulin-induced reduction in resistin mRNA (Fig. 4C), although increased p38 MAP-kinase activity by insulin was blocked with 10 µmol/l of SB203580 (Fig. 4D).
Next, 3T3-L1 adipocytes were preincubated with LY294002, an inhibitor of PI 3-kinase, for 30 min and incubated with or without 100 nmol/l insulin for 16 h with LY294002. LY294002 caused no change in basal resistin mRNA expression in cells unstimulated by insulin. Resistin mRNA expression in cells treated with insulin and LY294002 were about 50% lower (p <0.05) than those of the cells treated with insulin alone (Fig. 5A). To confirm the effect of LY294002, we measured the insulin-induced serine phosphorylation of Akt (also known as PKB), which is considered to be a downstream signal of PI 3-kinase pathway. Insulin-induced serine phosphorylation of Akt was blocked by 50 µmol/l LY294002 (Fig. 5B), suggesting that the PI 3-kinase activity was blocked by the inhibitor.
Effects of PI 3-kinase inhibition on insulin-induced decrease of resistin mRNA. (A) 3T3-L1 adipocytes were pretreated with or without LY294002 for 30 min and stimulated with insulin for 16 h. Resistin mRNA was examined by Northern blot analysis. Resistin mRNA was corrected with 36B4 mRNA. Data, expressed as percent of resistin mRNA in cells not treated with insulin or LY294002 (control), are shown as the means ± SEM of three independent experiments. *p<0.05 vs. control. †p<0.05 vs. LY294002-treated. #p<0.05 treatment with insulin vs. co-treatment with insulin and LY294002. (B) 3T3-L1 adipocytes were pretreated with or without LY294002 for 30 min and stimulated with insulin for 10 min. Total cell lysates were subjected to Western blot analysis with anti-phospho Akt antibody or anti-Akt antibody. (C, D) 3T3-L1 adipocytes were infected with AdexCALacZ or AdexCAΔp85. Resistin mRNA was examined by Northern blot analysis. Resistin mRNA was corrected with 36B4 mRNA. Data, expressed as percent of resistin mRNA in AdexCALacZ-infected cells without insulin (control), are shown as the means ± SEM of three independent experiments. *p<0.05 vs. control. †p<0.05 vs. AdexCAΔp85-infected. n.s.: no significant change. (C). PI 3-kinase assay was carried out. Data, expressed as percent of 32P-labelled PI in AdexCALacZ-infected cells with insulin (control), are shown as the means ± SEM of three independent experiments
To investigate the effect of PI 3-kinase pathway in detail, 3T3-L1 adipocytes were overexpressed with a dominant negative form of PI 3-kinase (Δp85). Although the insulin-induced PI 3-kinase activity was notably reduced (20% of control cells) in the cells overexpressed with Δp85, there was no difference in resistin mRNA expression between cells expressing LacZ and Δp85 after insulin treatment (Fig. 5C,D).
The above results indicate that insulin-induced down-regulation of resistin mRNA is mediated via the ERK1/2, p38 MAP-kinase or PI 3-kinase independent pathways.
To investigate whether insulin-induced down-regulation of resistin mRNA expression was mediated via synthesis of other protein(s), 3T3-L1 adipocytes were pretreated with cycloheximide, a protein synthesis inhibitor, for 30 min and stimulated with or without insulin for 16 h with cycloheximide. Treatment with cycloheximide alone did not cause any change in basal resistin mRNA level (Fig. 6A,B). Importantly, the decrease of resistin mRNA by insulin was completely blocked by co-treatment with cycloheximide, suggesting the effect of insulin on resistin mRNA requires the synthesis of new protein(s).
Effect of cycloheximide (CHX) on insulin-induced decrease of resistin mRNA. (A) 3T3-L1 adipocytes were pretreated with or without CHX (5 µg/ml) and stimulated with insulin for 16 h. Resistin mRNA was examined by Northern blot analysis. Equal load of the total RNA was confirmed by 28S/18S ribosomal RNA. (B) Data, expressed as percent of resistin mRNA in cells not treated with insulin or CHX (control), are expressed as the means ± SEM of three independent experiments. *p<0.05 vs. control. #p<0.05 treatment with insulin vs. co-treatment with insulin and CHX
We then treated the cells with actinomycin D, a RNA synthesis inhibitor, to examine the half-life of resistin mRNA (Fig. 7A,B). The calculated half-life of resistin mRNA was 12.7 h in 3T3-L1 adipocytes treated with actinomycin D (Fig. 7B, open circles). Interestingly, the decreasing rate of resistin mRNA with insulin was clearly faster than that of actinomycin D alone (Fig. 7B, open triangles), indicating that insulin accelerates the degradation of resistin mRNA. This effect of insulin was completely blocked by the co-treatment of cells with actinomycin D (Fig. 7B, open squares), and the resistin mRNA expression in cells with the co-treatment were comparable with those in cells treated with actinomycin D alone at any time points (Fig. 7A,B). This was probably because actinomycin D blocked the transcription of both the resistin gene and other gene(s) that encoded the protein(s) responsible for the degradation of resistin mRNA. Considered together with the results of cycloheximide, these findings suggested that insulin-induced decrease of resistin mRNA is mediated by the synthesis of mRNA of other gene(s) whose product could, at least in part, accelerate resistin mRNA degradation (Fig. 8).
Effect of actinomycin D (acti-D) on insulin-induced decrease of resistin mRNA. (A) 3T3-L1 adipocytes were pretreated with or without acti-D (5 µg/ml) and then stimulated with insulin for 4, 8 and 16 h. Resistin mRNA was examined by Northern blot analysis. Equal load of the total RNA was confirmed by 28S/18S ribosomal RNA. (B) Resistin mRNA was expressed as a percent of resistin mRNA in cells not treated with insulin or acti-D (control). (open circles; treatment with acti-D, open triangles; treatment with insulin, open squares; treatment with acti-D and insulin). Data are shown as the means ± SEM of three independent experiments
Proposed mechanism of insulin-induced decrease of resistin mRNA expression in 3T3-L1 adipocytes. Insulin stimulates the gene expression of unknown protein(s). The protein(s) could accelerate the degradation of resistin mRNA. It was not confirmed if the unknown protein(s) directly suppresses the transcription of resistin gene
Discussion
It is well known that insulin controls the expression of a number of genes involved in the metabolism of glucose or other nutrients [15]. Insulin is considered to be a positive regulator of resistin expression, since resistin expression decreases during fasting and increases after re-feeding with parallel changes in serum resistin concentration [6]. Indeed, administration of insulin in diabetic animals causes a marked increase in resistin mRNA [7]. In contrast, it has been reported that resistin mRNA is decreased in genetically obese mice (e.g. ob/ob mouse), which have supra-normal concentration of plasma insulin [8, 21]. Moreover, recent report has shown that insulin suppresses resistin mRNA expression in 3T3-L1 adipocytes [16]. To clarify the impact of insulin on resistin expression, we examined in this study the effects of insulin on resistin mRNA expression, and investigated the molecular mechanisms of such effect in 3T3-L1 cells.
In agreement with the results of another study [16], our results showed that insulin treatment reduced resistin mRNA expression in differentiated 3T3-L1 adipocytes. The change in resistin mRNA expression was significant for 8 h or longer treatment with insulin, and was insulin dose-dependent.
We then investigated the signalling pathways through which insulin could mediate the reduction of resistin mRNA. It is well known that PI 3-kinase pathway and MAP-kinase pathway are the two major signalling pathways activated by insulin [15]. To investigate the role of these pathways in the regulation of resistin mRNA expression, we separately used chemical inhibitors of each pathway. We found that neither MAP-kinase inhibitors (PD98059 for ERK1/2 and SB203580 for p38 MAP-kinase), nor the PI 3-kinase inhibitor prevented insulin-induced reduction of resistin mRNA. Another PI 3-kinase inhibitor, a dominant negative form of p85, also failed to block insulin-induced reduction of resistin mRNA. It is unlikely that the failure of prevention by these inhibitors is due to insufficient inhibition of these pathways, since we have confirmed the effects of these inhibitors in the cells.
Several PI 3-kinase and MAP-kinase independent pathways have been described in 3T3-L1 cells. The proto-oncogene product of c-Cbl is prominently tyrosine phosphorylated in response to insulin in 3T3-L1 adipocytes [22, 23]. It has been reported that CAP (c-Cbl-associating protein)-Cbl complex can modulate insulin-stimulated glucose transport and glycogen synthesis independent of PI 3-kinase or MAP-kinase [24]. It also has been reported that the action of insulin on gene expression involves both direct and indirect effects mediated through changes in glucose metabolism [25]. Therefore, we investigated whether insulin could reduce resistin mRNA expression in 3T3-L1 adipocytes cultured in glucose-free medium. Our results show that insulin-induced decrease of resistin mRNA occurred even in glucose-free medium, suggesting that the effect of insulin is not mediated through increased glucose metabolism.
In addition to the MAP-kinase and PI 3-kinase pathways, the JAK-STAT (signal transducers and activators of transcription) and JNK (c-Jun N-terminal protein kinase) pathways might be involved in the regulation of gene expression [26, 27, 28]. STAT 1, 3, 5 and 6 are reported to be expressed in 3T3-L1 adipocytes, however, insulin does not induce the translocation of those STATs from the cytosol to nucleus in 3T3-L1 adipocytes [29], and JNK is not phosphorylated by insulin [30]. Therefore, JAK-STAT and JNK pathways could not be related to insulin-induced decrease of resistin mRNA expression in 3T3-L1 adipocytes. Further studies are necessary to identify the signalling pathway that links insulin to resistin mRNA expression.
Cycloheximide, a protein synthesis inhibitor, completely blocked insulin-induced reduction of resistin mRNA. This strongly suggests that the effects of insulin are mediated through the synthesis of protein(s). The fact that the decrease of resistin mRNA occurred 8 h after insulin stimulation also supports the results that the effect of insulin is mediated though the synthesis of novel protein(s), because the direct regulation of gene transcription by insulin could occur within a shorter time (within 1 h in PEPCK gene [31]).
It is generally accepted that the regulation of gene expression by insulin occurs by controlling the transcription of genes or by changing the stability of the mRNA [15]. At the moment, the mechanism(s) by which insulin alters mRNA stability is entirely unknown. In this study, we have found that the resistin mRNA expression with insulin alone were clearly lower than those with actinomycin D alone. If insulin simply suppresses the transcription of reisistin gene and if it does not accelarate the degradation speed of resistin mRNA, resistin mRNA expression in cells treated with insulin alone should never be lower than those with actinomycin D alone. Therefore, we assume that insulin accelerates the degradation of resistin mRNA. Importantly, the decreasing rate of resistin mRNA in cells co-treated with insulin and actinomycin D is comparable with that in cells treated with actinomycin D alone, again suggesting that the effect of insulin is mediated via the synthesis of some other mRNA (and protein). Considering the results of cycloheximide and actinomycin D together, Fig. 8 provides a schematic representation of the proposed mechanisms of insulin-induced down-regulation of resistin mRNA. Insulin stimulates the expression of mRNA of unknown gene(s) via the ERK1/2, p38 MAP-kinase or PI 3-kinase independent pathways. The product(s) of the gene(s) could accelerate the degradation of resistin mRNA. It is not confirmed if the unknown protein(s) could suppress the transcription of resistin gene, since actinomycin D could also inhibit the synthesis of mRNA (and protein) of the unknown gene(s).
Resistin is a small peptide composed of 114 amino acids (murine) and is synthesized in adipose cells [6, 7, 32]. It has been shown that insulin down-regulated resistin protein content in differentiated 3T3-L1 adipocytes [33]. Our data and the previous studies suggest that insulin could reduce resistin protein content in 3T3-L1 adipocytes by suppressing the mRNA expression and the protein synthesis of resistin. It has been shown that resistin was secreted from adipose tissue and 3T3-L1 adipocytes after processing of the N-terminal signal peptides, however, the impact of insulin on these processes is unknown [6]. It has been reported that insulin also down-regulates the expression of several proteins by accelerating their degradation [34]. Therefore, it is important to examine the role of insulin on the secretion and the degradation of resistin protein.
In conclusion, we have shown that insulin down-regulated resistin mRNA via PI 3-kinase-, ERK- or p38 MAP-kinase-independent pathway in 3T3-L1 adipocytes. The decrease of resistin mRNA occurred through the synthesis of protein(s), which could accelerate the resistin mRNA degradation. As resistin might be a key molecule that links obesity to insulin resistance, further studies on the regulation of resistin expression should enhance our understanding of the mechanism(s) involved in the development of insulin resistance and may allow us to design new approaches to the treatment of Type 2 diabetes.
Abbreviations
- PI 3-kinase:
-
phosphatidylinositol 3-kinase
- ERK:
-
extracellular signal-regulated kinase
- MAP-kinase:
-
mitogen-activated protein-kinase
References
Saltiel AR (2000) The molecular and physiological basis of insulin resistance: emerging implications for metabolic and cardiovascular diseases. J Clin Invest 106:163–164
Kahn BB, Flier JS (2000) Obesity and insulin resistance. J Clin Invest 106:473–481
Hotamisligil GS, Shargill NS, Spiegelman BM (1993) Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science 259:87–91
Shimomura I, Hammer RE, Ikemoto S, Brown MS, Goldstein JL (1999) Leptin reverses insulin resistance and diabetes mellitus in mice with congenital lipodystrophy. Nature 401:73–76
Yamauchi T, Kamon J, Waki H et al. (2001) The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat Med 7:941–946
Steppan CM, Bailey ST, Bhat S et al. (2001) The hormone resistin links obesity to diabetes. Nature 409:307–312
Kim KH, Lee K, Moon YS, Sul HS (2001) A Cysteine-rich adipose tissue-specific secretory factor inhibits adipocyte differentiation. J Biol Chem 276:11252–11256
Way JM, Görgün CZ, Tong Q et al. (2001) Adipose tissue resistin expression is severely suppressed in obesity and stimulated by PPARγ agonists. J Biol Chem 276:25651–25653
Savage DB, Sewter CP, Klenk ES et al. (2001) Resistin/Fizz3 expression in relation to obesity and peroxisome proliferator-activated receptor-γ action in humans. Diabetes 50:2199–2202
Engert JC, Vohl MC, Williams SM et al. (2002) 5' flanking variants of resistin are associated with obesity. Diabetes 51:1629–1634
Nagaev I, Smith U (2001) Insulin resistance and type 2 diabetes are not related to resistin expression in human fat cells or skeletal muscle. Biochem Biophys Res Commun 285:561–564
Janke J, Engeli S, Gorzelniak K, Luft FC, Sharma AM (2002) Resistin gene expression in human adipocytes is not related to insulin resistance. Obes Res 10:1-5
Fasshauer M, Klein J, Neumann S, Eszlinger M, Paschke R (2001) Isoproterenol inhibits resistin gene expression through a Gs-protein-coupled pathway in 3T3-L1 adipocytes. FEBS Lett 500:60–63
Fasshauer M, Klein J, Neumann S, Eszlinger M, Paschke R (2001) Tumor necrosis factor alpha is a negative regulator of resistin gene expression and secretion in 3T3-L1 adipocytes. Biochem Biophys Res Commun 288:1027–1031
O'Brien RH, Granner DK (1996) Regulation of gene expression by insulin. Physiol Rev 76:1109–1161
Haugen F, Jørgensen A, Drevon CH, Trayhurn P (2001) Inhibition by insulin of resistin gene expression in 3T3-L1 adipocytes. FEBS Lett 507:105–108
Hunt CR, Ro JH, Dobson DE, Min HY, Spiegelman BM (1986) Adipocyte P2 gene: developmental expression and homology of 5'-flanking sequences among fat cell-specific genes. Proc Natl Acad Sci USA 83:3786–3790
Simpson DA, Feeney S, Boyle C, Stitt AW (2000) Retinal VEGF mRNA measured by SYBR Green I fluorescence: A versatile approach to quantitative PCR. Mol Vis 6:178–183
Tsuruzoe K, Emkey R, Kriauciunas KM, Ueki K, Kahn CR (2001) Insulin receptor substrate 3 (IRS-3) and IRS-4 impair IRS-1- and IRS-2-mediated signaling. Mol Cell Biol 21:26–38
Sakaue H, Ogawa W, Takata M et al. (1997) Phosphoinositide 3-kinase is required for insulin-induced but not for growth hormone- or hyperosmolarity-induced glucose uptake in 3T3-L1 adipocytes. Mol Endocrinol 11:1552–1562
Rajala MW, Lin Y, Ranalletta M et al. (2002) Cell type-specific expression and coregulation of murine resistin and resistin-like molecule-α in adipose tissue. Mol Endocrinol 16:1920–1930
Ribon V, Saltiel AR (1997) Insulin stimulates tyrosine phosphorylation of the proto-oncogene product of c-Cbl in 3T3-L1 adipocytes. Biochem J 324:839–846
Ribon V, Printen JA, Hoffman NG, Kay BK, Saltiel AR (1998) A novel, multifunctional c-Cbl binding protein in insulin receptor signaling in 3T3-L1 adipocytes. Mol Cell Biol 18:872–879
Baumann CA, Ribon V, Kanzaki M et al. (2000) CAP defines second signaling pathway required for insulin-stimulated glucose transport. Nature 407:202–207
Vaulont S, Kahn A (1994) Transcriptional control of metabolic regulation genes by carbohydrates. FASEB J 8:28–35
Coffer PJ, Puijenbroek A van, Burgering BMT et al. (1997) Insulin activates Stat3 independently of p21ras-ERK and PI-3 K signal transduction. Oncogene 15:2529–2539
Pierreux CE, Ursø B, Meyts PD, Rousseau GG, Lemaigre FP (1998) Inhibition by insulin of glucocorticoid-induced gene transcription: involvement of the ligand-binding domain of the glucocorticoid receptor and independence from the phosphatidylinositol 3-kinase and mitogen-activated protein kinase pathways. Mol Endocrinol 12:1344–1354
De Los Pinos E, Fernandez De Mattos S, Joaquin M, Tauler A (2001) Insulin inhibits glucocorticoid-stimulated L-type 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase gene expression by activation of the c-Jun N-terminal kinase pathway. Biochem J 353:267–273
Balhoff JP, Stephens JM (1998) Highly specific and quantitative activation of STATs in 3T3-L1 adipocytes. Biochem Biophys Res Commun 247:894–900
Kayali AG, Austin DA, Webster NJ (2000) Stimulation of MAPK cascades by insulin and osmotic shock: lack of an involvement of p38 mitogen-activated protein kinase in glucose transport in 3T3-L1 adipocytes. Diabetes 49:1783–1793
Sasaki K, Cripe TP, Koch SR et al. (1984) Multihormonal regulation of phosphoenolpyruvate carboxykinase gene transcription. The dominant role of insulin. J Biol Chem 259:15242–15251
Steppan CM, Brown EJ, Wright CM et al. (2001) A family of tissue-specific resistin-like molecules. Proc Natl Acad Sci USA 98:502–506
Shojima N, Sakoda H, Ogihara T et al. (2002) Humoral regulation of resistin expression in 3T3-L1 and mouse adipose cells. Diabetes 51:1737–1744
Zhande R, Mitchell JJ, Wu J, Sun XJ (2002) Molecular mechanism of insulin-induced degradation of insulin receptor substrate 1. Mol Cell Biol 22:1016–1026
Acknowledgements
We thank Dr. M. Kasuga (Kobe University, Kobe, Japan) for providing AdexCAΔp85 and Dr. K. Ueki (Joslin Diabetes Center, Boston, Mass., USA) for providing AdexCALacZ.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kawashima, J., Tsuruzoe, K., Motoshima, H. et al. Insulin down-regulates resistin mRNA through the synthesis of protein(s) that could accelerate the degradation of resistin mRNA in 3T3-L1 adipocytes. Diabetologia 46, 231–240 (2003). https://doi.org/10.1007/s00125-002-1022-3
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-002-1022-3