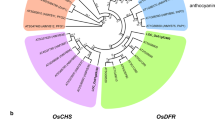
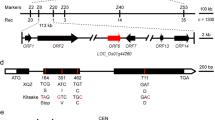
Abstract
Key message
we identified a functional chromogen gene C from wild rice, providing a new insight of anthocyanin biosynthesis pathway in indica and japonica.
Abstract
Accumulation of anthocyanin is a desirable trait to be selected in rice domestication, but the molecular mechanism of anthocyanin biosynthesis in rice remains largely unknown. In this study, a novel allele of chromogen gene C, OrC1, from Oryza rufipongon was cloned and identified as a determinant regulator of anthocyanin biosynthesis. Although OrC1 functions in purple apiculus, leaf sheath and stigma in indica background, it only promotes purple apiculus in japonica. Transcriptome analysis revealed that OrC1 regulates flavonoid biosynthesis pathway and activates a few bHLH and WD40 genes of ternary MYB-bHLH-WD40 complex in indica. Differentially expressed genes and metabolites were found in the indica and japonica backgrounds, indicating that OrC1 activated the anthocyanin biosynthetic genes OsCHI, OsF3H and OsANS and produced six metabolites independently. Artificial selection and domestication of C1 gene in rice occurred on the coding region in the two subspecies independently. Our results reveal the regulatory system and domestication of C1, provide new insights into MYB transcript factor involved in anthocyanin biosynthesis, and show the potential of engineering anthocyanin biosynthesis in rice.








Similar content being viewed by others
References
Aizza LCB, Dornelas MC (2011) A genomic approach to study anthocyanin synthesis and flower pigmentation in passionflowers. J Nucleic Acids 37:15–17
Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39:366–380
Chagne D, Lin-Wang K, Espley RV, Volz RK, How NM, Rouse S, Brendolise C, Carlisle CM, Kumar S, De Silva N, Micheletti D, McGhie T, Crowhurst RN, Storey RD, Velasco R, Hellens RP, Gardiner SE, Allan AC (2013) An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol 161:225–239
Chen W, Gong L, Guo Z, Wang WS, Zhang HY, Liu XQ, Yu SB (2013) A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol Plant 6:1769–1780
Cho YG, Ishii T, Temnykh S, Chen X, Lipovich L, McCouch SR, Park WD, Ayres N, Cartinhour S (2000) Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza sativa L.). Theor Appl Genet 100:713–722
Choi J, Platts A, Fuller D, Hsing Y, Wing R, Purugganan M (2017) The rice paradox: multiple origins but single domestication in Asian rice. Mol Biol Evol 34:969–979
Chin H, Wu Y, Hour A, Hong C, Lin Y (2016) Genetic and evolutionary analysis of purple leaf sheath in rice. Rice 9:8
de Vetten N, Quattrocchio F, Mol J, Koes, (1997) The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev 11:1422–1434
Fan FJ, Fan YY, Du JH (2008) Fine mapping of C (chromogen for anthocyanin) gene in rice. Rice Sci 15:1–6
Furukawa T, Maekawa M, Oki T, Suda I, Iida S, Shimada H, Takamure I, Kadowaki K (2007) The Rc and Rd genes are involved in pro-anthocyanidin synthesis in rice pericarp. Plant J 49:91–102
Gao DY, He B, Zhou YH, Sun LH (2011) Genetic and molecular analysis of a purple sheath somaclonal mutant in japonica rice. Plant Cell Rep 30:901–911
Grotewold E (2006) The genetics and biochemistry of floral pigments. Ann Rev Plant Biol 57:761–780
Hugueney P, Provenzano S, Verriès C, Ferrandino A, Meudec E, Batelli G, Merdinoglu D, Cheynier V, Schubert A, Ageorges A (2009) A novel cation-dependent O-methyltransferase involved in anthocyanin methylation in grapevine. Plant Physiol 150:2057–2070
Honda C, Kotoda N, Wada M, Kondo S, Kobayashi S, Soejima J, Zhang Z, Tsuda T, Moriguchi T (2002) Anthocyanin biosynthetic genes are coordinately expressed during red coloration in apple skin. Plant Physiol Biochem 40:955–962
Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V (2011) Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot 62:2465–2483
Hu W, Zhou T, Han Z, Tan C, Xing Y (2020) Dominant complementary interaction between OsC1 and two tightly linked genes, Rb1 and Rb2, controls the purple leaf sheath in rice. Theor Appl Genet 133:2555–2566
Huang X, Kurata N, Wei X, Wang Z, Wang A, Zhao Q, Zhao Y, Liu K, Lu H, Li W, Guo Y, Lu Y, Zhou C, Fan D, Weng Q, Zhu C, Huang T, Zhang L, Wang Y, Feng L, Furuumi H, Kubo T, Miyabayashi T, Yuan X, Xu Q, Dong G, Zhan Q, Li C, Fujiyama A, Toyoda A, Lu T, Feng Q, Qian Q, Li J, Han B (2012) A map of rice genome variation reveals the origin of cultivated rice. Nature 490:497–501
Jian W, Cao H, Yuan S, Liu Y, Lu J, Lu W, Li N, Wang J, Zou J, Tang N, Xu C, Cheng Y, Gao Y, Xi W, Bouzayen M, Li Z (2019) SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic Res 6:22
Jin W, Wang H, Li M, Wang J, Yang Y, Zhang X (2016) The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol J 14:2120–2133
Jun JH, Liu C, Xiao X, Dixon RA (2015) The transcriptional repressor MYB2 regulates both spatial and temporal patterns of proanthocyandin and anthocyanin pigmentation in Medicago truncatula. Plant Cell 27:2860–2879
Kanehisa M, Goto S (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30
Kim D, Pertea G, Trapnell C, Pimentel H, Ryan K, Salzberg S (2013) TopHat2: accurat e alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:36
Li CB, Zhou A, Sang T (2006) Rice domestication by reducing shattering. Science 311:1936–1939
Liu X, Sun X, Wang W, Ding H, Liu W, Li G, Jiang M, Zhu C, Yao F (2012) Fine mapping of Pa-6 gene for purple apiculus in rice. Plant Biotechnol J 55:218–225
Li J, Xu R, Wang C, Qi L, Zheng XM, Wang WS, Ding YB, Zhang LZ, Wang YY, Cheng YL, Zhang LF, Qiao WH, Yang QW (2018) A heading date QTL, qHD7.2, from wild rice (Oryza rufipogon) delays flowering and shortens panicle length under long-day conditions. Sci Rep 8:2928
Livak K, Schmittgen T (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-[Δ][Δ]Ct method. Methods 25:402–408
Mikami I, Takahashi A, Khin-Thidar SY (2000) A candidate for C (chromogen for anthocyanin) gene. Rice Genet Newsl 17:54–56
Qi L, Ding YB, Zheng XM, Xu R, Zhang LZ, Wang YY, Wang XN, Zhang LF, Cheng YL, Qiao WH, Yang QW (2018) Fine mapping and identification of a novel locus qGL12.2 controls grain length in wild rice (Oryza rufipogon Griff.). Theor Appl Genet 131:1497–1508
Qiao WH, Qi L, Cheng ZJ, Su L, Li J, Sun Y, Ren JF, Zheng XM, Yang QW (2016) Development and characterization of chromosome segment substitution lines derived from Oryza rufipogon in the genetic background of O. sativa spp. indica cultivar 9311. BMC Genomics 17:580
Saitoh K, Onishi K, Mikami I, Thidar K, Sano Y (2004) Allelic diversification at the C (OsC1) locus of wild and cultivated rice: nucleotide changes associated with phenotypes. Genetics 168:997–1007
Sakamoto W, Ohmori T, Kageyama K, Miyazaki C, Saito A, Murata M, Noda K, Maekawa M (2001) The purple leaf (Pl) locus of rice: the Pl(W) allele has a complex organization and includes two genes encoding basic helix-loop-helix proteins involved in anthocyanin biosynthesis. Plant Cell Physiol 42:982–991
Sun X, Zhang Z, Chen C, Wu W, Ren N, Jiang C, Yu J, Zhao Y, Zheng X, Yang Q, Zhang H, Li J, Li Z (2018) The C-S-A gene system regulates hull pigmentation and reveals evolution of anthocyanin biosynthesis pathway in rice. J Exp Bot 69:1485–1498
Shin Y, Park H, Yim S, Baek N, Lee C, An G, Woo Y (2006) Transgenic rice lines expressing maize C1 and R-S regulatory genes produce various flavonoids in the endosperm. Plant Biotechnol J 4:303–315
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tan L, Li X, Liu F, Sun X, Li C, Zhu Z, Fu Y, Cai H, Wang X, Xie D, Sun C (2008) Control of a key transition from prostrate to erect growth in rice domestication. Nat Genet 40:1360–1364
Tian J, Peng Z, Zhang J, Song T, Wan H, Zhang M, Yao Y (2015) McMYB10 regulates coloration via activating McF3’H and later structural genes in ever-red leaf crabapple. Plant Biotechnol J 13:948–961
Vinayagam R, Xu B (2015) Antidiabetic properties of dietary flavonoids: a cellular mechanism review. Nutr Metab 12:60
Xu W, Dubos C, Lepiniec L (2015) Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci 20:176–185
Xu Z, Yang Q, Feng K, Yu X, Xiong A (2020) DcMYB113, a root-specific R2R3-MYB, conditions anthocyanin biosynthesis and modification in carrot. Plant Biotechnol J 18:1585–1597
Yuan H, Zeng X, Shi J, Xu Q, Wang Y, Jabu D, Sang Z, Nyima T (2018) Time-course comparative metabolite profiling under osmotic stress in tolerant and sensitive Tibetan hulless barley. BioMed Res Int 9415409:1–12
Zhang LZ, Huang JF, Wang YY, Xu R, Yang ZY, Zhao ZG, Liu SJ, Tian Y, Zheng XM, Li F, Wang JR, Song Y, Li JQ, Cui YX, Zhang LF, Cheng YL, Lan JH, Qiao WH, Yang QW (2020) Identification and genetic analysis of qCL1.2, a novel allele of the “green revolution” gene SD1 from wild rice (Oryza rufipogon) that enhances plant height. BMC Genet 21:62
Zhang Q, Wang L, Liu Z, Zhao Z, Zhao J, Wang Z, Zhou G, Liu P, Liu M (2020) Transcriptome and metabolome profiling unveil the mechanisms of Ziziphus jujuba Mill. peel coloration. Food Chem 312:125903
Zhang Y, Su JB, Duan S, Ao DJR, Liu J, Wang P, Li YG, Liu B, Feng DR, Wang JF, Wang HB (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7:30
Zhao SS, Wang CH, Ma J, Wang S, Tian P, Wang JL, Cheng ZJ, Zhang X, Guo XP, Lei CL (2016) Map-based cloning and functional analysis of the chromogen gene C in rice (Oryza sativa L.). J Plant Biol 59:496–505
Zheng J, Wu H, Zhu HB, Huang C, Liu C, Chang YS, Kong ZC, Zou ZH, Wang GW, Lin YJ, Chen H (2019) Determining factors, regulation system, and domestication of anthocyanin biosynthesis in rice leaves. New Phytologis 223:705–721
Zhu Z, Schultz AW, Wang J, Johnson CH, Yannone SM, Patti GJ, Siuzdak G (2013) Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat Protoc 8:451–456
Acknowledgements
We especially thank Dr. Peng Zhang (The University of Sydney, Australia) and Dr. Hao Chen (Huazhong Agricultural University, China) for critical reading of the manuscript. This work was supported by the National Key R&D Program of China (2016YFD0100101) and the National Natural Science Foundation of China (31471471).
Author information
Authors and Affiliations
Contributions
WH Qiao, JH Lan and QW Yang conceived and designed the experiments. WH Qiao and YY Wang performed the experiments and wrote the paper. R Xu, ZY Yang, JR Wang and JF Huang analyzed the data. Y Sun, L Su and LZ Zhang performed fine mapping. SJ Liu, YL Tian, LM Chen and X Liu contributed to field investigation. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Jessica Rutkoski.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
122_2021_3787_MOESM9_ESM.tif
Genotype analysis of promoter region of C1. (a) Sequence polymorphism of different haplotypes of C1 promoter from 180 accessions. No. of Acc.: number of accessions, I: O. sativa. indica, J: O. sativa. japonica, R: O. rufipongon, N: O. nivara. (b) Haplotype network of C1 promoter.
Supplementary file 8 (TIF 8402 kb)
122_2021_3787_MOESM10_ESM.tif
Genotype analysis of promoter region of C1. (a) Sequence polymorphism of different haplotypes of C1 promoter from 180 accessions. No. of Acc.: number of accessions, I: O. sativa. indica, J: O. sativa. japonica, R: O. rufipongon, N: O. nivara. (b) Haplotype network of C1 promoter.
Supplementary file 8 (TIF 2267 kb)
Rights and permissions
About this article
Cite this article
Qiao, W., Wang, Y., Xu, R. et al. A functional chromogen gene C from wild rice is involved in a different anthocyanin biosynthesis pathway in indica and japonica. Theor Appl Genet 134, 1531–1543 (2021). https://doi.org/10.1007/s00122-021-03787-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-021-03787-1