Abstract
Key message
Genomic selection using data from an on-going breeding program can improve gain from selection, relative to phenotypic selection, by significantly increasing the number of lines that can be evaluated.
Abstract
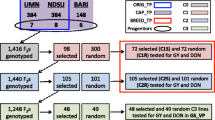
The early stages of phenotyping involve few observations and can be quite inaccurate. Genomic selection (GS) could improve selection accuracy and alter resource allocation. Our objectives were (1) to compare the prediction accuracy of GS and phenotyping in stage-1 and stage-2 field evaluations and (2) to assess the value of stage-1 phenotyping for advancing lines to stage-2 testing. We built training populations from 1769 wheat breeding lines that were genotyped and phenotyped for yield, test weight, Fusarium head blight resistance, heading date, and height. The lines were in cohorts, and analyses were done by cohort. Phenotypes or GS estimated breeding values were used to determine the trait value of stage-1 lines, and these values were correlated with their phenotypes from stage-2 trials. This was repeated for stage-2 to stage-3 trials. The prediction accuracy of GS and phenotypes was similar to each other regardless of the amount (0, 50, 100%) of stage-1 data incorporated in the GS model. Ranking of stage-1 lines by GS predictions that used no stage-1 phenotypic data had marginally lower correspondence to stage-2 phenotypic rankings than rankings of stage-1 lines based on phenotypes. Stage-1 lines ranked high by GS had slightly inferior phenotypes in stage-2 trials than lines ranked high by phenotypes. Cost analysis indicated that replacing stage-1 phenotyping with GS would allow nearly three times more stage-1 candidates to be assessed and provide 0.84–2.23 times greater gain from selection. We conclude that GS can complement or replace phenotyping in early stages of phenotyping.




Similar content being viewed by others
Abbreviations
- AST1:
-
Predictions and selections based on GEBVs using all stage-1 phenotypic data
- FHB:
-
Fusarium head blight
- FST1:
-
Predictions and selections based on GEBVs using ½ stage-1 phenotypic data-based selecting lines based on family relations
- GEBV:
-
Genomic estimated breeding values
- GS:
-
Genomic selection
- NST1:
-
Predictions and selections based on GEBVs using no stage-1 phenotypic data
- NST1-1K:
-
Same as NST1 except predictions made with a just 10% of the markers
- PHEN:
-
Predictions and selections based on phenotypes
- PS:
-
Phenotypic selection
- RST1:
-
Predictions and selections based on GEBVs using ½ stage-1 phenotypic data based on random selection of lines
- TP:
-
Training population
References
Abed A, Pérez-Rodríguez P, Crossa J, Belzile F (2018) When less can be better: how can we make genomic selection more cost-effective and accurate in barley? Theor Appl Genet 131(9):1873–1890
Appels R, International Wheat Genome Sequencing Consortium (IWGSC) et al (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:6403
Bassi FM, Bentley AR, Charmet G, Ortiz R, Crossa J (2015) Breeding schemes for the implementation of genomic selection in wheat (Triticum spp.). Plant Sci 242:23–36
Bates D, Mächler M, Bolker BM, Walker SC (2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67(1):1
Belamkar V, Guttieri MJ, Hussain W, Jarquín D, El-basyoni I, Poland J, Lorenz A, Baenziger PS (2018) Genomic selection in preliminary yield trials in a winter wheat breeding program. G3: Genes Genomes Genetics 8(8):2735–2747
Bernal-Vasquez A-M, Gordillo A, Schmidt M, Piepho H-P (2017) Genomic prediction in early selection stages using multi-year data in a hybrid rye breeding program. BMC Genet 18(1):51
Buckler E, Ilut D, Wang X, Kretzschmar T, Gore M, Mitchell S. (2016) rAmpSeq: Using repetitive sequences for robust genotyping. bioRxiv, 096628. https://doi.org/10.1101/096628
Burgueño J, de los Campos G, Weigel K, Crossa J (2012) Genomic prediction of breeding values when modeling genotype x environment interaction using pedigree and dense molecular markers. Crop Sci 52:707–719
Butler D, Cullis B, Gilmour A, Gogel B (2009) ASReml-R reference manual. The State of Queensland, Department of Primary Industries and Fisheries, Brisbane, Australia
Covarrubias-Pazaran G (2016) Genome assisted prediction of quantitative traits using the R package sommer. PLoS ONE 11(6):e0156744
Crossa J, Burgueno J, Cornelius P, McLaren G, Trethowan R, Krishnamachari A (2006) Modeling genotype x environment interaction using additive genetic covariances of relatives for predicting breeding values of wheat genotypes. Crop Sci 46:1722–1733
Endelman JB (2011) Ridge regression and other Kernels for genomic selection with R package rrBLUP. Plant Genome J 4(3):250
Garrick DJ, Taylor JF, Fernando RL (2009) Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet Sel Evol. https://doi.org/10.1186/1297-9686-41-55
Gaynor RC, Gorjanc G, Bentley A, Ober E, Howell P, Jackson R, MacKay I, Hickey J (2017) A two-part strategy for using genomic selection to develop inbred lines. Crop Sci 57(5):2372–2386
Glaubitz J, Casstevens T, Lu F, Harriman J, Elshire R, Sun Q, Buckler E (2014) TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS ONE. https://doi.org/10.1371/journal.pone.0090346
Goudet J, Jombart T (2015) hierfstat: estimation and tests of hierarchical F-statistics. R package version 0.04-22
Habier D, Fernando R, Dekkers JC (2007) The impact of genetic relationship information on genome-assisted breeding values. Genetics 177(4):2389–2397
Hartl DL, Clark GC (1997) Principles of population genetics. Sinauer Associates, Sunderland
He S, Schulthess A, Mirdita V, Zhao Y, Korzun V, Bothe B, Ebermeyer E, Reif J, Jiang Y (2016) Genomic selection in a commercial winter wheat population. Theor Appl Genetics 1293:641–651
Heffner EL, Lorenz AJ, Jannink J-L, Sorrells ME (2010) Plant breeding with genomic selection: gain per unit time and cost. Crop Sci 50:1681–1690. https://doi.org/10.2135/cropsci2009.11.0662
Heffner EL, Jannink J-L, Sorrells ME (2011a) Genomic selection accuracy using multifamily prediction models in a wheat breeding program. Plant Genome 4(1):65
Heffner EL, Jannink J-L, Iwata H, Sorrells E, Sorrells ME (2011b) Genomic selection accuracy for grain quality traits in biparental wheat populations. Crop Sci 51:2597–2606. https://doi.org/10.2135/cropsci2011.05.0253
Hoffstetter A, Cabrera A, Huang M, Sneller C (2016) Optimizing training population data and validation of genomic selection for economic traits in soft winter wheat. G3: Genes Genomes Genetics 6(9):2919–2928
Huang M, Cabrera A, Hoffstetter A, Griffey C, Van Sanford D, Costa J, McKendry A, Sneller C (2016) Genomic selection for wheat traits and trait stability. Theor Appl Genetics 129(9):1697–1710
Huang M, Ward B, Griffey C, Van Sanford D, McKendry A, Brown-Guedira G, Tyagi P, Sneller C (2018) The accuracy of genomic prediction between environments and populations for soft wheat traits. Crop Sci 58(6):2274–2288
Jannink J-L, Lorenz A, Iwata H (2010) Genomic selection in plant breeding: from theory to practice. Briefings Funct Genomics Proteom 9(2):166–177
Longin C, Mi X, Würschum T (2015) Genomic selection in wheat: optimum allocation of test resources and comparison of breeding strategies for line and hybrid breeding. Theor Appl Genetics 128(7):1297–1306
Lopez-Cruz M, Crossa J, Bonnett D, Dreisigacker S, Poland J, Jannink J-L, Singh RP, Autrique E, de los Campos G (2015) Increased prediction accuracy in wheat breeding trials using a marker × environment interaction genomic selection model. G3: Genes Genomes Genetics 5(4):569–582
Marulanda J, Mi X, Melchinger AE, Xu J-L, Würschum T, Longin C (2016) Optimum breeding strategies using genomic selection for hybrid breeding in wheat, maize, rye, barley, rice and triticale. Theor Appl Genetics 129(10):1901–1913
Meuwissen TH, Hayes B, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157(4):1819–1829
Michel S, Ametz C, Gungor H, Epure D, Grausgruber H, Löschenberger F, Buerstmayr H (2016) Genomic selection across multiple breeding cycles in applied bread wheat breeding. Theor Appl Genetics 129(6):1179–1189
Michel S, Ametz C, Gungor H, Akgöl B, Epure D, Grausgruber H, Löschenberger F, Buerstmayr H (2017) Genomic assisted selection for enhancing line breeding: merging genomic and phenotypic selection in winter wheat breeding programs with preliminary yield trials. Theor Appl Genetics 130(2):363–376
Nazarian A, Gezan S (2016) GenoMatrix: a software package for pedigree-based and genomic prediction analyses on complex traits. J Hered 107(4):372–379. https://doi.org/10.1093/jhered/esw020Epub 2016 Mar 29
Ostersen T, Christensen OF, Henryon M, Neilson B, Su G, Madsen P (2011) Deregressed EBV as the response variable yield more reliable genomic predictions than traditional EBV in pure-bred pigs. Genet Sel Evol. https://doi.org/10.1186/1297-9686-43-38
Pembleton L, Inch C, Baillie R, Drayton M, Thakur P, Ogaji Y, Spangenberg G, Forster JW, Daetwyler HD, Cogan N (2018) Exploitation of data from breeding programs supports rapid implementation of genomic selection for key agronomic traits in perennial ryegrass. Theor Appl Genetics 131(9):1891–1902
Philomi J, Montesinos-Lopez OA, Crossa J, Mondal S, Perez LG, Poland J, Huerta-Espino J, Crespo-Herrera L, Govindan V, Dreisigacker S, Shretha S, Perez-Rodriguez P, Espinosa FP, Singh RP (2019) Integrating genomic-enabled prediction and high-throughput phenotyping in breeding for climate-resilient bread wheat. Theor Appl Genet 132:177–194. https://doi.org/10.1007/s00122-018-3206-3
Piepho HP, Möhring J, Melchinger AE, Büchse A (2008) BLUP for phenotypic selection in plant breeding and variety testing. Euphytica 161(1–2):209–228
Poland J, Brown P, Sorrells M, Jannink J-L (2012) Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS ONE 7(2):32253
R Core Team (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Rajsic P, Weersink A, Navabi A, Peter Pauls K (2016) Economics of genomic selection: the role of prediction accuracy and relative genotyping costs. Euphytica 210(2):259–276
Rinaldo A, Bacanu S-A, Devlin B, Sonpar V, Wasserman L, Roeder K (2005) Characterization of multilocus linkage disequilibrium. Genetic Epidemiol 28(3):193–206
Rodríguez-Álvarez M, Boer M, van Eeuwijk FA, Eilers P (2018) Correcting for spatial heterogeneity in plant breeding experiments with P-splines. Spatial Stat 23:52–71
Rutkoski J, Poland J, Mondal S, Autrique E, Pérez L, Crossa J, Reynolds M, Singh R (2016) Canopy temperature and vegetation indices from high-throughput phenotyping improve accuracy of pedigree and genomic selection for grain yield in wheat. G3: Genes Genomes Genetics 6(9):2799–2808
Sallam AH, Smith KP (2016) Genomic selection performs similarly to phenotypic selection in barley. Crop Sci 56(6):2871–2881
SAS Institute (2017) Base SAS 9.4 procedures guide: Statistical procedures
Sneller C, Paul P, Guttieri MJ (2010) Characterization of resistance to Fusarium head blight in an eastern US soft red winter wheat population. Crop Sci 50(1):123–133
Song J, Carver B, Powers C, Yan L, Klapste L, El-Kassaby Y, Chen C (2017) Practical application of genomic selection in a double-haploid winter wheat breeding program. Mol Breed 37:117. https://doi.org/10.1007/x11032-017-0715-8
Sun J, Poland J, Mondal S, Crossa J, Juliana P, Singh R, Rutkoski J, Jannink J-L, Crespo-Herrera I, Velu G, Huerta-Espino H, Sorrells ME (2019) High-throughput phenotyping platforms enhance genomic selection for wheat grain yield across populations and cycles in early stage. Theor Appl Genetics 132(6):1705–1720
Tolhurst D, Mathews K, Smith A, Cullis B (2019) Genomic selection in multi-environment plant breeding trials using a factor analytic linear mixed model. J Anim Breed Genet 136:279–300
Velazco J, Rodríguez-Álvarez M, Boer M, Jordan D, Eilers P, Malosetti M, van Eeuwijk FA (2017) Modelling spatial trends in sorghum breeding field trials using a two-dimensional P-spline mixed model. Theor Appl Genetics 130(7):1375–1392
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38(6):1358–1370
Acknowledgements
We would like to thank Cassi Sewell, Duc Hua, Brian Sugerman, and all the employees of the OSU winter wheat breeding program that assisted in collecting the data used in this analysis. Without them this work would not have been possible, and for this, we are truly grateful. We acknowledge funding from Ohio Agricultural Research and Development Center, National Institute for Food and Agriculture, and the US Wheat and Barley Scab Initiative.
Funding
The funding was provided by Ohio Small Grains Marketing Program, Agricultural Research Service (Grant No. 1234567).
Author information
Authors and Affiliations
Contributions
DB executed the analyses and wrote the manuscript, MH assisted in the data analyses and editing the manuscript, EO executed the genotyping-by-sequencing, CS initiated the study, directed the project, performed some data analyses, and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Communicated by Jose Crossa.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix: Prediction accuracy by trait, stage, prediction method, and cohort
Appendix: Prediction accuracy by trait, stage, prediction method, and cohort
Trait | Cohort | Stages | Model | Prediction accuracy |
|---|---|---|---|---|
Yield | OH12 | 2013 Stage-1 to 2014 Stage-2 | PHEN | − 0.060 |
Yield | OH13 | 2014 Stage-1 to 2015 Stage-2 | PHEN | 0.098 |
Yield | OH12 | 2014 Stage-2 to 2015 Stage-3 | PHEN | 0.383 |
Yield | OH14 | 2015 Stage-1 to 2016 Stage-2 | PHEN | 0.027 |
Yield | OH13 | 2015 Stage-2 to 2016 Stage-3 | PHEN | − 0.015 |
Yield | OH15 | 2016 Stage-1 to 2017 Stage-2 | PHEN | 0.413 |
Yield | OH14 | 2016 Stage-2 to 2017 Stage-3 | PHEN | 0.302 |
Yield | OH16 | 2017 Stage-1 to 2018 Stage-2 | PHEN | 0.210 |
Yield | OH15 | 2017 Stage-2 to 2018 Stage-3 | PHEN | 0.517 |
Yield | OH12 | 2013 Stage-1 to 2014 Stage-2 | AST1 | 0.022 |
Yield | OH13 | 2014 Stage-1 to 2015 Stage-2 | AST1 | 0.130 |
Yield | OH12 | 2014 Stage-2 to 2015 Stage-3 | AST1 | 0.309 |
Yield | OH14 | 2015 Stage-1 to 2016 Stage-2 | AST1 | 0.008 |
Yield | OH13 | 2015 Stage-2 to 2016 Stage-3 | AST1 | 0.009 |
Yield | OH15 | 2016 Stage-1 to 2017 Stage-2 | AST1 | 0.450 |
Yield | OH14 | 2016 Stage-2 to 2017 Stage-3 | AST1 | 0.416 |
Yield | OH16 | 2017 Stage-1 to 2018 Stage-2 | AST1 | 0.246 |
Yield | OH15 | 2017 Stage-2 to 2018 Stage-3 | AST1 | 0.528 |
Yield | OH12 | 2013 Stage-1 to 2014 Stage-2 | NST1 | 0.171 |
Yield | OH13 | 2014 Stage-1 to 2015 Stage-2 | NST1 | 0.181 |
Yield | OH12 | 2014 Stage-2 to 2015 Stage-3 | NST1 | 0.456 |
Yield | OH14 | 2015 Stage-1 to 2016 Stage-2 | NST1 | 0.127 |
Yield | OH13 | 2015 Stage-2 to 2016 Stage-3 | NST1 | − 0.039 |
Yield | OH15 | 2016 Stage-1 to 2017 Stage-2 | NST1 | 0.219 |
Yield | OH14 | 2016 Stage-2 to 2017 Stage-3 | NST1 | 0.444 |
Yield | OH16 | 2017 Stage-1 to 2018 Stage-2 | NST1 | 0.127 |
Yield | OH15 | 2017 Stage-2 to 2018 Stage-3 | NST1 | 0.536 |
Yield | OH12 | 2013 Stage-1 to 2014 Stage-2 | RST1 | 0.053 |
Yield | OH13 | 2014 Stage-1 to 2015 Stage-2 | RST1 | 0.108 |
Yield | OH14 | 2015 Stage-1 to 2016 Stage-2 | RST1 | 0.010 |
Yield | OH15 | 2016 Stage-1 to 2017 Stage-2 | RST1 | 0.360 |
Yield | OH16 | 2017 Stage-1 to 2018 Stage-2 | RST1 | 0.272 |
Yield | OH12 | 2013 Stage-1 to 2014 Stage-2 | FST1 | 0.053 |
Yield | OH13 | 2014 Stage-1 to 2015 Stage-2 | FST1 | 0.108 |
Yield | OH14 | 2015 Stage-1 to 2016 Stage-2 | FST1 | 0.015 |
Yield | OH15 | 2016 Stage-1 to 2017 Stage-2 | FST1 | 0.356 |
Yield | OH16 | 2017 Stage-1 to 2018 Stage-2 | FST1 | 0.266 |
Yield | OH12 | 2013 Stage-1 to 2014 Stage-2 | NST1-1K | 0.267 |
Yield | OH13 | 2014 Stage-1 to 2015 Stage-2 | NST1-1K | 0.163 |
Yield | OH12 | 2014 Stage-2 to 2015 Stage-3 | NST1-1K | 0.429 |
Yield | OH14 | 2015 Stage-1 to 2016 Stage-2 | NST1-1K | 0.141 |
Yield | OH13 | 2015 Stage-2 to 2016 Stage-3 | NST1-1K | 0.026 |
Yield | OH15 | 2016 Stage-1 to 2017 Stage-2 | NST1-1K | 0.071 |
Yield | OH14 | 2016 Stage-2 to 2017 Stage-3 | NST1-1K | 0.296 |
Yield | OH16 | 2017 Stage-1 to 2018 Stage-2 | NST1-1K | 0.074 |
Yield | OH15 | 2017 Stage-2 to 2018 Stage-3 | NST1-1K | 0.490 |
Yield | OH12 | 2013 Stage-1 to 2014 Stage-2 | NST1-0.5 | 0.147 |
Yield | OH13 | 2014 Stage-1 to 2015 Stage-2 | NST1-0.5 | 0.039 |
Yield | OH12 | 2014 Stage-2 to 2015 Stage-3 | NST1-0.5 | 0.418 |
Yield | OH14 | 2015 Stage-1 to 2016 Stage-2 | NST1-0.5 | 0.104 |
Yield | OH13 | 2015 Stage-2 to 2016 Stage-3 | NST1-0.5 | − 0.077 |
Yield | OH15 | 2016 Stage-1 to 2017 Stage-2 | NST1-0.5 | 0.190 |
Yield | OH14 | 2016 Stage-2 to 2017 Stage-3 | NST1-0.5 | 0.339 |
Yield | OH16 | 2017 Stage-1 to 2018 Stage-2 | NST1-0.5 | 0.088 |
Yield | OH15 | 2017 Stage-2 to 2018 Stage-3 | NST1-0.5 | 0.542 |
Yield | OH12 | 2013 Stage-1 to 2014 Stage-2 | NST1-1.5 | 0.126 |
Yield | OH13 | 2014 Stage-1 to 2015 Stage-2 | NST1-1.5 | 0.112 |
Yield | OH12 | 2014 Stage-2 to 2015 Stage-3 | NST1-1.5 | 0.416 |
Yield | OH14 | 2015 Stage-1 to 2016 Stage-2 | NST1-1.5 | 0.126 |
Yield | OH13 | 2015 Stage-2 to 2016 Stage-3 | NST1-1.5 | − 0.058 |
Yield | OH15 | 2016 Stage-1 to 2017 Stage-2 | NST1-1.5 | 0.225 |
Yield | OH14 | 2016 Stage-2 to 2017 Stage-3 | NST1-1.5 | 0.419 |
Yield | OH16 | 2017 Stage-1 to 2018 Stage-2 | NST1-1.5 | 0.088 |
Yield | OH15 | 2017 Stage-2 to 2018 Stage-3 | NST1-1.5 | 0.529 |
Test Weight | OH12 | 2014 Stage-2 to 2015 Stage-3 | PHEN | 0.352 |
Test Weight | OH13 | 2015 Stage-2 to 2016 Stage-3 | PHEN | 0.477 |
Test Weight | OH14 | 2016 Stage-2 to 2017 Stage-3 | PHEN | 0.558 |
Test Weight | OH15 | 2017 Stage-2 to 2018 Stage-3 | PHEN | − 0.170 |
Test Weight | OH12 | 2014 Stage-2 to 2015 Stage-3 | AST1 | 0.141 |
Test Weight | OH13 | 2015 Stage-2 to 2016 Stage-3 | AST1 | 0.316 |
Test Weight | OH14 | 2016 Stage-2 to 2017 Stage-3 | AST1 | 0.583 |
Test Weight | OH15 | 2017 Stage-2 to 2018 Stage-3 | AST1 | − 0.178 |
Test Weight | OH12 | 2014 Stage-2 to 2015 Stage-3 | NST1 | 0.157 |
Test Weight | OH13 | 2015 Stage-2 to 2016 Stage-3 | NST1 | 0.338 |
Test Weight | OH14 | 2016 Stage-2 to 2017 Stage-3 | NST1 | 0.469 |
Test Weight | OH15 | 2017 Stage-2 to 2018 Stage-3 | NST1 | − 0.145 |
Height | OH12 | 2013 Stage-1 to 2014 Stage-2 | PHEN | 0.572 |
Height | OH13 | 2014 Stage-1 to 2015 Stage-2 | PHEN | 0.285 |
Height | OH12 | 2014 Stage-2 to 2015 Stage-3 | PHEN | 0.395 |
Height | OH14 | 2015 Stage-1 to 2016 Stage-2 | PHEN | |
Height | OH13 | 2015 Stage-2 to 2016 Stage-3 | PHEN | 0.472 |
Height | OH15 | 2016 Stage-1 to 2017 Stage-2 | PHEN | 0.520 |
Height | OH14 | 2016 Stage-2 to 2017 Stage-3 | PHEN | 0.488 |
Height | OH16 | 2017 Stage-1 to 2018 Stage-2 | PHEN | 0.376 |
Height | OH15 | 2017 Stage-2 to 2018 Stage-3 | PHEN | 0.646 |
Height | OH12 | 2013 Stage-1 to 2014 Stage-2 | AST1 | 0.281 |
Height | OH13 | 2014 Stage-1 to 2015 Stage-2 | AST1 | 0.312 |
Height | OH12 | 2014 Stage-2 to 2015 Stage-3 | AST1 | 0.321 |
Height | OH14 | 2015 Stage-1 to 2016 Stage-2 | AST1 | |
Height | OH13 | 2015 Stage-2 to 2016 Stage-3 | AST1 | 0.436 |
Height | OH15 | 2016 Stage-1 to 2017 Stage-2 | AST1 | 0.494 |
Height | OH14 | 2016 Stage-2 to 2017 Stage-3 | AST1 | 0.409 |
Height | OH16 | 2017 Stage-1 to 2018 Stage-2 | AST1 | 0.283 |
Height | OH15 | 2017 Stage-2 to 2018 Stage-3 | AST1 | 0.499 |
Height | OH12 | 2013 Stage-1 to 2014 Stage-2 | NST1 | 0.057 |
Height | OH13 | 2014 Stage-1 to 2015 Stage-2 | NST1 | 0.281 |
Height | OH12 | 2014 Stage-2 to 2015 Stage-3 | NST1 | 0.353 |
Height | OH14 | 2015 Stage-1 to 2016 Stage-2 | NST1 | |
Height | OH13 | 2015 Stage-2 to 2016 Stage-3 | NST1 | 0.288 |
Height | OH15 | 2016 Stage-1 to 2017 Stage-2 | NST1 | 0.042 |
Height | OH14 | 2016 Stage-2 to 2017 Stage-3 | NST1 | 0.492 |
Height | OH16 | 2017 Stage-1 to 2018 Stage-2 | NST1 | 0.209 |
Height | OH15 | 2017 Stage-2 to 2018 Stage-3 | NST1 | 0.439 |
Height | OH12 | 2013 Stage-1 to 2014 Stage-2 | RST1 | 0.433 |
Height | OH13 | 2014 Stage-1 to 2015 Stage-2 | RST1 | 0.350 |
Height | OH14 | 2015 Stage-1 to 2016 Stage-2 | RST1 | − 0.015 |
Height | OH15 | 2016 Stage-1 to 2017 Stage-2 | RST1 | 0.353 |
Height | OH16 | 2017 Stage-1 to 2018 Stage-2 | RST1 | 0.228 |
Height | OH12 | 2013 Stage-1 to 2014 Stage-2 | FST1 | 0.433 |
Height | OH13 | 2014 Stage-1 to 2015 Stage-2 | FST1 | 0.350 |
Height | OH14 | 2015 Stage-1 to 2016 Stage-2 | FST1 | − 0.015 |
Height | OH15 | 2016 Stage-1 to 2017 Stage-2 | FST1 | 0.356 |
Height | OH16 | 2017 Stage-1 to 2018 Stage-2 | FST1 | 0.225 |
Heading Date | OH12 | 2013 Stage-1 to 2014 Stage-2 | PHEN | 0.434 |
Heading Date | OH13 | 2014 Stage-1 to 2015 Stage-2 | PHEN | 0.686 |
Heading Date | OH12 | 2014 Stage-2 to 2015 Stage-3 | PHEN | 0.664 |
Heading Date | OH14 | 2015 Stage-1 to 2016 Stage-2 | PHEN | 0.588 |
Heading Date | OH13 | 2015 Stage-2 to 2016 Stage-3 | PHEN | 0.726 |
Heading Date | OH15 | 2016 Stage-1 to 2017 Stage-2 | PHEN | 0.607 |
Heading Date | OH14 | 2016 Stage-2 to 2017 Stage-3 | PHEN | 0.752 |
Heading Date | OH16 | 2017 Stage-1 to 2018 Stage-2 | PHEN | 0.619 |
Heading Date | OH15 | 2017 Stage-2 to 2018 Stage-3 | PHEN | 0.744 |
Heading Date | OH12 | 2013 Stage-1 to 2014 Stage-2 | AST1 | 0.471 |
Heading Date | OH13 | 2014 Stage-1 to 2015 Stage-2 | AST1 | 0.660 |
Heading Date | OH12 | 2014 Stage-2 to 2015 Stage-3 | AST1 | 0.614 |
Heading Date | OH14 | 2015 Stage-1 to 2016 Stage-2 | AST1 | 0.564 |
Heading Date | OH13 | 2015 Stage-2 to 2016 Stage-3 | AST1 | 0.681 |
Heading Date | OH15 | 2016 Stage-1 to 2017 Stage-2 | AST1 | 0.621 |
Heading Date | OH14 | 2016 Stage-2 to 2017 Stage-3 | AST1 | 0.720 |
Heading Date | OH16 | 2017 Stage-1 to 2018 Stage-2 | AST1 | 0.645 |
Heading Date | OH15 | 2017 Stage-2 to 2018 Stage-3 | AST1 | 0.734 |
Heading Date | OH12 | 2013 Stage-1 to 2014 Stage-2 | NST1 | 0.282 |
Heading Date | OH13 | 2014 Stage-1 to 2015 Stage-2 | NST1 | 0.576 |
Heading Date | OH12 | 2014 Stage-2 to 2015 Stage-3 | NST1 | 0.670 |
Heading Date | OH14 | 2015 Stage-1 to 2016 Stage-2 | NST1 | 0.191 |
Heading Date | OH13 | 2015 Stage-2 to 2016 Stage-3 | NST1 | 0.643 |
Heading Date | OH15 | 2016 Stage-1 to 2017 Stage-2 | NST1 | 0.284 |
Heading Date | OH14 | 2016 Stage-2 to 2017 Stage-3 | NST1 | 0.707 |
Heading Date | OH16 | 2017 Stage-1 to 2018 Stage-2 | NST1 | 0.502 |
Heading Date | OH15 | 2017 Stage-2 to 2018 Stage-3 | NST1 | 0.774 |
Heading Date | OH12 | 2013 Stage-1 to 2014 Stage-2 | RST1 | 0.496 |
Heading Date | OH13 | 2014 Stage-1 to 2015 Stage-2 | RST1 | 0.698 |
Heading Date | OH14 | 2015 Stage-1 to 2016 Stage-2 | RST1 | 0.418 |
Heading Date | OH15 | 2016 Stage-1 to 2017 Stage-2 | RST1 | 0.519 |
Heading Date | OH16 | 2017 Stage-1 to 2018 Stage-2 | RST1 | 0.559 |
Heading Date | OH12 | 2013 Stage-1 to 2014 Stage-2 | FST1 | 0.496 |
Heading Date | OH13 | 2014 Stage-1 to 2015 Stage-2 | FST1 | 0.698 |
Heading Date | OH14 | 2015 Stage-1 to 2016 Stage-2 | FST1 | 0.435 |
Heading Date | OH15 | 2016 Stage-1 to 2017 Stage-2 | FST1 | 0.516 |
Heading Date | OH16 | 2017 Stage-1 to 2018 Stage-2 | FST1 | 0.573 |
FHB Index | OH12 | 2013 Stage-1 to 2014 Stage-2 | PHEN | 0.155 |
FHB Index | OH13 | 2014 Stage-1 to 2015 Stage-2 | PHEN | |
FHB Index | OH12 | 2014 Stage-2 to 2015 Stage-3 | PHEN | 0.402 |
FHB Index | OH14 | 2015 Stage-1 to 2016 Stage-2 | PHEN | 0.027 |
FHB Index | OH13 | 2015 Stage-2 to 2016 Stage-3 | PHEN | 0.543 |
FHB Index | OH15 | 2016 Stage-1 to 2017 Stage-2 | PHEN | 0.494 |
FHB Index | OH14 | 2016 Stage-2 to 2017 Stage-3 | PHEN | 0.454 |
FHB Index | OH16 | 2017 Stage-1 to 2018 Stage-2 | PHEN | 0.566 |
FHB Index | OH15 | 2017 Stage-2 to 2018 Stage-3 | PHEN | 0.556 |
FHB Index | OH12 | 2013 Stage-1 to 2014 Stage-2 | AST1 | 0.280 |
FHB Index | OH13 | 2014 Stage-1 to 2015 Stage-2 | AST1 | |
FHB Index | OH12 | 2014 Stage-2 to 2015 Stage-3 | AST1 | 0.343 |
FHB Index | OH14 | 2015 Stage-1 to 2016 Stage-2 | AST1 | 0.066 |
FHB Index | OH13 | 2015 Stage-2 to 2016 Stage-3 | AST1 | 0.408 |
FHB Index | OH15 | 2016 Stage-1 to 2017 Stage-2 | AST1 | 0.536 |
FHB Index | OH14 | 2016 Stage-2 to 2017 Stage-3 | AST1 | 0.448 |
FHB Index | OH16 | 2017 Stage-1 to 2018 Stage-2 | AST1 | 0.588 |
FHB Index | OH15 | 2017 Stage-2 to 2018 Stage-3 | AST1 | 0.604 |
FHB Index | OH12 | 2013 Stage-1 to 2014 Stage-2 | NST1 | 0.271 |
FHB Index | OH13 | 2014 Stage-1 to 2015 Stage-2 | NST1 | |
FHB Index | OH12 | 2014 Stage-2 to 2015 Stage-3 | NST1 | 0.197 |
FHB Index | OH14 | 2015 Stage-1 to 2016 Stage-2 | NST1 | 0.280 |
FHB Index | OH13 | 2015 Stage-2 to 2016 Stage-3 | NST1 | 0.444 |
FHB Index | OH15 | 2016 Stage-1 to 2017 Stage-2 | NST1 | 0.282 |
FHB Index | OH14 | 2016 Stage-2 to 2017 Stage-3 | NST1 | 0.512 |
FHB Index | OH16 | 2017 Stage-1 to 2018 Stage-2 | NST1 | 0.356 |
FHB Index | OH15 | 2017 Stage-2 to 2018 Stage-3 | NST1 | 0.530 |
FHB Index | OH12 | 2013 Stage-1 to 2014 Stage-2 | RST1 | 0.230 |
FHB Index | OH13 | 2014 Stage-1 to 2015 Stage-2 | RST1 | 0.382 |
FHB Index | OH14 | 2015 Stage-1 to 2016 Stage-2 | RST1 | 0.065 |
FHB Index | OH15 | 2016 Stage-1 to 2017 Stage-2 | RST1 | 0.463 |
FHB Index | OH16 | 2017 Stage-1 to 2018 Stage-2 | RST1 | 0.525 |
FHB Index | OH12 | 2013 Stage-1 to 2014 Stage-2 | FST1 | 0.230 |
FHB Index | OH13 | 2014 Stage-1 to 2015 Stage-2 | FST1 | 0.382 |
FHB Index | OH14 | 2015 Stage-1 to 2016 Stage-2 | FST1 | 0.065 |
FHB Index | OH15 | 2016 Stage-1 to 2017 Stage-2 | FST1 | 0.444 |
FHB Index | OH16 | 2017 Stage-1 to 2018 Stage-2 | FST1 | 0.534 |
FHB Index | OH12 | 2013 Stage-1 to 2014 Stage-2 | 1RPHEN | 0.169 |
FHB Index | OH13 | 2014 Stage-1 to 2015 Stage-2 | 1RPHEN | 0.473 |
FHB Index | OH14 | 2015 Stage-1 to 2016 Stage-2 | 1RPHEN | 0.030 |
FHB Index | OH15 | 2016 Stage-1 to 2017 Stage-2 | 1RPHEN | 0.401 |
FHB Index | OH16 | 2017 Stage-1 to 2018 Stage-2 | 1RPHEN | 0.497 |
FHB Index | OH12 | 2013 Stage-1 to 2014 Stage-2 | 1RST1 | 0.276 |
FHB Index | OH13 | 2014 Stage-1 to 2015 Stage-2 | 1RST1 | 0.407 |
FHB Index | OH14 | 2015 Stage-1 to 2016 Stage-2 | 1RST1 | 0.052 |
FHB Index | OH15 | 2016 Stage-1 to 2017 Stage-2 | 1RST1 | 0.442 |
FHB Index | OH16 | 2017 Stage-1 to 2018 Stage-2 | 1RST1 | 0.555 |
FHB Index | OH12 | 2013 Stage-1 to 2014 Stage-2 | NST1-1K | 0.234 |
FHB Index | OH13 | 2014 Stage-1 to 2015 Stage-2 | NST1-1K | 0.237 |
FHB Index | OH12 | 2014 Stage-2 to 2015 Stage-3 | NST1-1K | 0.166 |
FHB Index | OH14 | 2015 Stage-1 to 2016 Stage-2 | NST1-1K | 0.232 |
FHB Index | OH13 | 2015 Stage-2 to 2016 Stage-3 | NST1-1K | 0.491 |
FHB Index | OH15 | 2016 Stage-1 to 2017 Stage-2 | NST1-1K | 0.285 |
FHB Index | OH14 | 2016 Stage-2 to 2017 Stage-3 | NST1-1K | 0.488 |
FHB Index | OH16 | 2017 Stage-1 to 2018 Stage-2 | NST1-1K | 0.262 |
FHB Index | OH15 | 2017 Stage-2 to 2018 Stage-3 | NST1-1K | 0.480 |
FHB Index | OH12 | 2013 Stage-1 to 2014 Stage-2 | NST1-0.5 | 0.237 |
FHB Index | OH13 | 2014 Stage-1 to 2015 Stage-2 | NST1-0.5 | − 0.058 |
FHB Index | OH12 | 2014 Stage-2 to 2015 Stage-3 | NST1-0.5 | 0.184 |
FHB Index | OH14 | 2015 Stage-1 to 2016 Stage-2 | NST1-0.5 | 0.317 |
FHB Index | OH13 | 2015 Stage-2 to 2016 Stage-3 | NST1-0.5 | 0.299 |
FHB Index | OH15 | 2016 Stage-1 to 2017 Stage-2 | NST1-0.5 | 0.249 |
FHB Index | OH14 | 2016 Stage-2 to 2017 Stage-3 | NST1-0.5 | 0.457 |
FHB Index | OH16 | 2017 Stage-1 to 2018 Stage-2 | NST1-0.5 | 0.264 |
FHB Index | OH15 | 2017 Stage-2 to 2018 Stage-3 | NST1-0.5 | 0.512 |
FHB Index | OH12 | 2013 Stage-1 to 2014 Stage-2 | NST1-1.5 | 0.281 |
FHB Index | OH13 | 2014 Stage-1 to 2015 Stage-2 | NST1-1.5 | 0.063 |
FHB Index | OH12 | 2014 Stage-2 to 2015 Stage-3 | NST1-1.5 | 0.189 |
FHB Index | OH14 | 2015 Stage-1 to 2016 Stage-2 | NST1-1.5 | 0.278 |
FHB Index | OH13 | 2015 Stage-2 to 2016 Stage-3 | NST1-1.5 | 0.299 |
FHB Index | OH15 | 2016 Stage-1 to 2017 Stage-2 | NST1-1.5 | 0.282 |
FHB Index | OH14 | 2016 Stage-2 to 2017 Stage-3 | NST1-1.5 | 0.501 |
FHB Index | OH16 | 2017 Stage-1 to 2018 Stage-2 | NST1-1.5 | 0.326 |
FHB Index | OH15 | 2017 Stage-2 to 2018 Stage-3 | NST1-1.5 | 0.520 |
Rights and permissions
About this article
Cite this article
Borrenpohl, D., Huang, M., Olson, E. et al. The value of early-stage phenotyping for wheat breeding in the age of genomic selection. Theor Appl Genet 133, 2499–2520 (2020). https://doi.org/10.1007/s00122-020-03613-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-020-03613-0