Abstract
Key message
Association mapping of resistance to Pyrenophora teres f. teres in a collection of Nordic barley germplasm at different developmental stages revealed 13 quantitative loci with mostly small effects.
Abstract
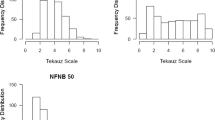
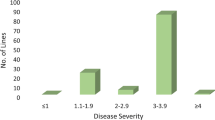
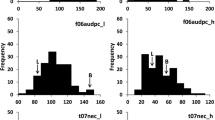
Net blotch, caused by the necrotrophic fungus Pyrenophora teres, is one of the major diseases in barley in Norway causing quantitative and qualitative yield losses. Resistance in Norwegian cultivars and germplasm is generally insufficient and resistance sources have not been extensively explored yet. In this study, we mapped quantitative trait loci (QTL) associated with resistance to net blotch in Nordic germplasm. We evaluated a collection of 209 mostly Nordic spring barley lines for reactions to net form net blotch (NFNB; Pyrenophora teres f. teres) in inoculations with three single conidia isolates at the seedling stage and in inoculated field trials at the adult stage in 4 years. Using 5669 SNP markers genotyped with the Illumina iSelect 9k Barley SNP Chip and a mixed linear model accounting for population structure and kinship, we found a total of 35 significant marker-trait associations for net blotch resistance, corresponding to 13 QTL, on all chromosomes. Out of these QTL, seven conferred resistance only in adult plants and four were only detectable in seedlings. Two QTL on chromosomes 3H and 6H were significant during both seedling inoculations and adult stage field trials. These are promising candidates for breeding programs using marker-assisted selection strategies. The results elucidate the genetic background of NFNB resistance in Nordic germplasm and suggest that NB resistance is conferred by a number of genes each with small-to-moderate effects, making it necessary to pyramid these genes to achieve sufficient levels of resistance.









Similar content being viewed by others
References
Abu Qamar M, Liu Z, Faris J, Chao S, Edwards M, Lai Z, Franckowiak J, Friesen T (2008) A region of barley chromosome 6H harbors multiple major genes associated with net type net blotch resistance. Theor Appl Genet 117:1261–1270
AHDB (2013) Fungicide activity and performance in barley. The Agriculture and Horticulture Development Board (AHDB) Information sheet 27, winter 2013/14. https://cereals.ahdb.org.uk/media/305416/is27-fungicide-activity-and-performance-in-barley.pdf. Accessed 23 June 2017
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265
Begum H, Spindel JE, Lalusin A, Borromeo T, Gregorio G, Hernandez J, Virk P, Collard B, McCouch SR (2015) Genome-wide association mapping for yield and other agronomic traits in an elite breeding population of tropical rice (Oryza sativa). PLoS One 10:e0119873
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Burlakoti R, Gyawali S, Chao S, Smith K, Horsley R, Cooper B, Muehlbauer G, Neate S (2016) Genome-wide association study of spot form of net blotch resistance in the upper midwest barley breeding programs. Phytopathology 107:100–108
Cakir M, Gupta S, Platz G, Ablett GA, Loughman R, Emebiri L, Poulsen D, Li C, Lance R, Galwey N (2003) Mapping and validation of the genes for resistance to Pyrenophora teres f. teres in barley (Hordeum vulgare L.). Crop Pasture Sci 54:1369–1377
Caldwell KS, Russell J, Langridge P, Powell W (2006) Extreme population-dependent linkage disequilibrium detected in an inbreeding plant species, Hordeum vulgare. Genetics 172:557–567
Cantalapiedra CP, Boudiar R, Casas AM, Igartua E, Contreras-Moreira B (2015) BARLEYMAP: physical and genetic mapping of nucleotide sequences and annotation of surrounding loci in barley. Mol Breed 35:1–11
Chan EK, Rowe HC, Kliebenstein DJ (2010) Understanding the evolution of defense metabolites in Arabidopsis thaliana using genome-wide association mapping. Genetics 185:991–1007
Chono M, Honda I, Zeniya H, Yoneyama K, Saisho D, Takeda K, Takatsuto S, Hoshino T, Watanabe Y (2003) A semidwarf phenotype of barley uzu results from a nucleotide substitution in the gene encoding a putative brassinosteroid receptor. Plant Physiol 133:1209–1219
Core Team R (2016) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Ellwood SR, Syme RA, Moffat CS, Oliver RP (2012) Evolution of three Pyrenophora cereal pathogens: recent divergence, speciation and evolution of non-coding DNA. Fungal Genet Biol 49:825–829
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Franckowiak J, Lundqvist U (1997) BGS 6, six-rowed spike 1, vrs1. Barley Genet Newsl 26:49–50
Friesen T, Faris J, Lai Z, Steffenson B (2006) Identification and chromosomal location of major genes for resistance to Pyrenophora teres in a doubled-haploid barley population. Genome 49:855–859
Goddard R, Peraldi A, Ridout C, Nicholson P (2014) Enhanced disease resistance caused by BRI1 mutation is conserved between Brachypodium distachyon and barley (Hordeum vulgare). Mol Plant Microbe Interact 27:1095–1106
Grewal T, Rossnagel B, Pozniak C, Scoles G (2008) Mapping quantitative trait loci associated with barley net blotch resistance. Theor Appl Genet 116:529–539
Grewal TS, Rossnagel BG, Scoles GJ (2012) Mapping quantitative trait loci associated with spot blotch and net blotch resistance in a doubled-haploid barley population. Mol Breed 30:267–279
Gupta S, Li C, Loughman R, Cakir M, Platz G, Westcott S, Bradley J, Broughton S, Lance R (2010) Quantitative trait loci and epistatic interactions in barley conferring resistance to net type net blotch (Pyrenophora teres f. teres) isolates. Plant Breed 129:362–368
Gupta S, Li C, Loughman R, Cakir M, Westcott S, Lance R (2011) Identifying genetic complexity of 6H locus in barley conferring resistance to Pyrenophora teres f. teres. Plant Breed 130:423–429
Gupta PK, Kulwal PL, Jaiswal V (2014) Association mapping in crop plants: opportunities and challenges. Adv Genet 85:109–147
Gutiérrez L, Germán S, Pereyra S, Hayes PM, Pérez CA, Capettini F, Locatelli A, Berberian NM, Falconi EE, Estrada R (2015) Multi-environment multi-QTL association mapping identifies disease resistance QTL in barley germplasm from Latin America. Theor Appl Genet 128:501–516
Hill W, Weir B (1988) Variances and covariances of squared linkage disequilibria in finite populations. Theor Popul Biol 33:54–78
Holland JB (2007) Genetic architecture of complex traits in plants. Curr Opin Plant Biol 10:156–161
Koladia V, Faris J, Richards J, Brueggeman R, Chao S, Friesen T (2016) Genetic analysis of net form net blotch resistance in barley lines CIho 5791 and Tifang against a global collection of P. teres f. teres isolates. Theor Appl Genet 130:163–173
König J, Perovic D, Kopahnke D, Ordon F (2013) Development of an efficient method for assessing resistance to the net type of net blotch (Pyrenophora teres f. teres) in winter barley and mapping of quantitative trait loci for resistance. Mol Breed 32:641–650
Liu Z, Ellwood SR, Oliver RP, Friesen TL (2011) Pyrenophora teres: profile of an increasingly damaging barley pathogen. Mol Plant Pathol 12:1–19
Liu Z, Holmes DJ, Faris JD, Chao S, Brueggeman RS, Edwards MC, Friesen TL (2015) Necrotrophic effector-triggered susceptibility (NETS) underlies the barley–Pyrenophora teres f. teres interaction specific to chromosome 6H. Mol Plant Pathol 16:188–200
Lundqvist U, Franckowiack J (1997) BGS 315, Six-rowed spike 3, vrs3. Barley Genet Newsl 26:264–265
Mascher M, Muehlbauer GJ, Rokhsar DS, Chapman J, Schmutz J, Barry K, Muñoz-Amatriaín M, Close TJ, Wise RP, Schulman AH (2013) Anchoring and ordering NGS contig assemblies by population sequencing (POPSEQ). Plant J 76:718–727
Massman J, Cooper B, Horsley R, Neate S, Dill-Macky R, Chao S, Dong Y, Schwarz P, Muehlbauer G, Smith K (2011) Genome-wide association mapping of Fusarium head blight resistance in contemporary barley breeding germplasm. Mol Breed 27:439–454
Mathre D (1997) Compendium of barley diseases, 2nd edn. American Phytopathological Society, St. Paul
Matthies IE, Malosetti M, Röder MS, van Eeuwijk F (2014) Genome-wide association mapping for kernel and malting quality traits using historical European barley records. PLoS One 9:e110046
McLean MS, Howlett BJ, Hollaway GJ (2009) Epidemiology and control of spot form of net blotch (Pyrenophora teres f. maculata) of barley: a review. Crop Pasture Sci 60:303–315
Muñoz-Amatriaín M, Cuesta-Marcos A, Endelman JB, Comadran J, Bonman JM, Bockelman HE, Chao S, Russell J, Waugh R, Hayes PM (2014) The USDA barley core collection: genetic diversity, population structure, and potential for genome-wide association studies. PLoS One 9:e94688
Nordborg M, Borevitz JO, Bergelson J, Berry CC, Chory J, Hagenblad J, Kreitman M, Maloof JN, Noyes T, Oefner PJ (2002) The extent of linkage disequilibrium in Arabidopsis thaliana. Nat Genet 30:190–193
Pasam RK, Sharma R, Malosetti M, van Eeuwijk FA, Haseneyer G, Kilian B, Graner A (2012) Genome-wide association studies for agronomical traits in a world wide spring barley collection. BMC Plant Biol 12:1
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Richards J, Chao S, Friesen T, Brueggeman R (2016) Fine mapping of the barley chromosome 6H net form net blotch susceptibility locus. G3 (Bethesda) 6:1809–1818
Richards JK, Friesen TL, Brueggeman RS (2017) Association mapping utilizing diverse barley lines reveals net form net blotch seedling resistance/susceptibility loci. Theor Appl Genet 130:915–927
Robinson J (2000) Yield of doubled haploid lines of Nordic spring barley infected with net blotch, Pyrenophora teres. Plant Breed 119:219–222
Robinson J, Jalli M (1999) Sensitivity of resistance to net blotch in barley. J Phytopathol 147:235–241
Roy JK, Smith KP, Muehlbauer GJ, Chao S, Close TJ, Steffenson BJ (2010) Association mapping of spot blotch resistance in wild barley. Mol Breed 26:243–256
Smedegård-Petersen V (1971) Pyrenophora teres f. maculata f. nov. and Pyrenophora teres f. teres on barley in Denmark. Yearb R Vet Agric Univ (Copenhagen) 1971:124–144
Somers D, Manninen O, Jalli M, Kalendar R, Schulman A, Afanasenko O, Robinson J (2006) Mapping of major spot-type and net-type net-blotch resistance genes in the Ethiopian barley line CI 9819. Genome 49:1564–1571
Statistics Norway (2016) Area used for grain and oil seed (table 04607). https://www.ssb.no/en/jord-skog-jakt-og-fiskeri/statistikker/korn. Accessed 23 June 2017
Steffenson B, Hayes P, Kleinhofs A (1996) Genetics of seedling and adult plant resistance to net blotch (Pyrenophora teres f. teres) and spot blotch (Cochliobolus sativus) in barley. Theor Appl Genet 92:552–558
Stracke S, Haseneyer G, Veyrieras J-B, Geiger HH, Sauer S, Graner A, Piepho H-P (2009) Association mapping reveals gene action and interactions in the determination of flowering time in barley. Theor Appl Genet 118:259–273
Tamang P, Neupane A, Mamidi S, Friesen T, Brueggeman R (2015) Association mapping of seedling resistance to spot form net blotch in a worldwide collection of barley. Phytopathology 105:500–508
Tekauz A (1985) A numerical scale to classify reactions of barley to Pyrenophora teres. Can J Plant Pathol 7:181–183
Wang M, Jiang N, Jia T, Leach L, Cockram J, Waugh R, Ramsay L, Thomas B, Luo Z (2012) Genome-wide association mapping of agronomic and morphologic traits in highly structured populations of barley cultivars. Theor Appl Genet 124:233–246
Wang X, Mace ES, Platz GJ, Hunt CH, Hickey LT, Franckowiak JD, Jordan DR (2015) Spot form of net blotch resistance in barley is under complex genetic control. Theor Appl Genet 128:489–499
Williams KJ, Smyl C, Lichon A, Wong KY, Wallwork H (2001) Development and use of an assay based on the polymerase chain reaction that differentiates the pathogens using spot form and net form of net blotch of barley. Australas Plant Pathol 30:37–44
Wonneberger R, Ficke A, Lillemo M (2017) Mapping of quantitative trait loci associated with resistance to net form net blotch (Pyrenophora teres f. teres) in a doubled haploid Norwegian barley population. PLoS One 12:e0175773
Zhu C, Gore M, Buckler ES, Yu J (2008) Status and prospects of association mapping in plants. Plant Genome 1:5–20
Acknowledgements
This work was supported by funding from the Research Council of Norway (NFR; Project 224833). The Nordic Barley Panel was developed in collaboration with Graminor and kind contributions from barley breeders Lars Reitan and Stein Bergersen at Graminor and Åsmund Bjørnstad and Helge Skinnes at NMBU. We also thank Timothy Friesen at USDA-ARS in Fargo, USA, for teaching seedling inoculation and scoring methodology, and Anja Karine Ruud and all laboratory and field technicians involved in this work for their contributions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest in the reported research.
Additional information
Communicated by Kevin Smith.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wonneberger, R., Ficke, A. & Lillemo, M. Identification of quantitative trait loci associated with resistance to net form net blotch in a collection of Nordic barley germplasm. Theor Appl Genet 130, 2025–2043 (2017). https://doi.org/10.1007/s00122-017-2940-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-017-2940-2