Abstract
Key message
This manuscript provides a Brassica conserved ortholog set (COS) that can be used as diagnostic cross-species markers as well as tools for genetic mapping and genome comparison of the Brassicaceae.
Abstract
A conserved ortholog set (COS) is a collection of genes that are conserved in both sequence and copy number between closely related genomes. COS is a useful resource for developing gene-based markers and is suitable for comparative genome mapping. We developed a COS for Brassica based on proteome comparisons of Arabidopsis thaliana, B. rapa, and B. oleracea to establish a basis for comparative genome analysis of crop species in the Brassicaceae. A total of 1,194 conserved orthologous single-copy genes were identified from the genomes based on whole-genome BLASTP analysis. Gene ontology analysis showed that most of them encoded proteins with unknown function and chloroplast-related genes were enriched. In addition, 152 Brassica COS primer sets were applied to 16 crop and wild species of the Brassicaceae and 57.9–92.8 % of them were successfully amplified across the species representing that a Brassica COS can be used as diagnostic cross-species markers of diverse Brassica species. We constructed a genetic map of Raphanus sativus by analyzing the segregation of 322 COS genes in an F2 population (93 individuals) of Korean cultivars (WK10039 × WK10024). Comparative genome analysis based on the COS genes showed conserved genome structures between R. sativus and B. rapa with lineage-specific rearrangement and fractionation of triplicated subgenome blocks indicating close evolutionary relationship and differentiation of the genomes. The Brassica COS developed in this study will play an important role in genetic, genomic, and breeding studies of crop Brassicaceae species.






Similar content being viewed by others
References
Al-Shehbaz I, Beilstein M, Kellogg E (2006) Systematics and phylogeny of the Brassicaceae (Cruciferae): an overview. Plant Syst Evol 259:89–120
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Arias T, Beilstein M, Tang M, McKain M, Pires J (2014) Diversification times among Brassica (Brassicaceae) crops suggest hybrid formation after 20 million years of divergence. Am J Bot 101:86–91
Beilstein M, Al-Shehbaz I, Kellogg E (2006) Brassicaceae phylogeny and trichome evolution. Am J Bot 93:607–619
Beilstein M, Nagalingum N, Clements M, Manchester S, Mathews S (2010) Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc Natl Acad Sci USA 107:18724–18728
Bushakra JM, Sargent DJ, Cabrera A, Crowhurst R, Girona EL, Velasco R, Symonds VV, Knaap E, Troggio M, Gardiner SE, Chagné D (2011) Rosaceae conserved orthologous set (RosCOS) markers as a tool to assess genome synteny between Malus and Fragaria. Tree Genet Genomes 8:643–658
Cabrera A, Kozik A, Howad W, Arus P, Iezzoni AF, van der Knaap E (2009) Development and bin mapping of a Rosaceae conserved ortholog set (COS) of markers. BMC Genom 10:562
Chapman MA, Chang J, Weisman D, Kesseli RV, Burke JM (2007) Universal markers for comparative mapping and phylogenetic analysis in the Asteraceae (Compositae). Theor Appl Genet 115:747–755
Cheng F, Wu J, Fang L, Wang X (2012) Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Front Plant Sci 3:198
Cheng F, Mandakova T, Wu J, Xie Q, Lysak MA, Wang X (2013) Deciphering the diploid ancestral genome of the Mesohexaploid Brassica rapa. Plant Cell 25:1541–1554
Dassanayake M, Oh DH, Haas JS, Hernandez A, Hong H, Ali S, Yun DJ, Bressan RA, Zhu JK, Bohnert HJ, Cheeseman JM (2011) The genome of the extremophile crucifer Thellungiella parvula. Nat Genet 43:913–918
Economic Research Service USDA (2008) Vegetables and melons outlook. http://www.ers.usda.gov/Publications/VGS/Tables/World.pdf
Fulton TM, Van der Hoeven R, Eannetta NT, Tanksley SD (2002) Identification, analysis, and utilization of conserved ortholog set markers for comparative genomics in higher plants. Plant Cell 14:1457–1467
Goldman N, Yang Z (1994) A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol 11:725–736
Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, Haberer G, Hollister JD, Ossowski S, Ottilar RP, Salamov AA, Schneeberger K, Spannagl M, Wang X, Yang L, Nasrallah ME, Bergelson J, Carrington JC, Gaut BS, Schmutz J, Mayer KF, Van de Peer Y, Grigoriev IV, Nordborg M, Weigel D, Guo YL (2011) The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet 43:476–481
Huang S, Deng L, Guan M, Li J, Lu K, Wang H, Fu D, Mason A, Liu S, Hua W (2013) Identification of genome-wide single nucleotide polymorphisms in allopolyploid crop Brassica napus. BMC Genom 14:717
Kim B, Yu H, Park S, Shin J, Oh M, Kim N, Mun J (2012) Identification and profiling of novel microRNAs in the Brassica rapa genome based on small RNA deep sequencing. BMC Plant Biol 12:218
Krutovsky KV, Troggio M, Brown GR, Jermstad KD, Neale DB (2004) Comparative mapping in the Pinaceae. Genetics 168:447–461
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Li F, Hasegawa Y, Saito M, Shirasawa S, Fukushima A, Ito T, Fujii H, Kishitani S, Kitashiba H, Nishio T (2011) Extensive chromosome homoeology among Brassiceae species were revealed by comparative genetic mapping with high-density EST-based SNP markers in radish (Raphanus sativus L.). DNA Res 18:401–411
Liewlaksaneeyanawin C, Zhuang J, Tang M, Farzaneh N, Lueng G, Cullis C, Findlay S, Ritland CE, Bohlmann J, Ritland K (2008) Identification of COS markers in the Pinaceae. Tree Genet Genomes 5:247–255
Lü N, Yamane K, Ohnishi O (2008) Genetic diversity of cultivated and wild radish and phylogenetic relationships among Raphanus and Brassica species revealed by the analysis of trnK/matK sequence. Breed Sci 58:15–22
Lysak M, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I (2006) Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc Natl Acad Sci USA 103:5224–5229
Mun J-H, Kwon S, Yang T, Seol Y, Jin M, Kim J, Lim M, Kim J, Baek S, Choi B, Yu H, Kim D, Kim N, Lim K, Lee S, Hahn J, Lim Y, Bancroft I, Park B (2009) Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol 10:R111
Nelson MN, Parkin IA, Lydiate DJ (2011) The mosaic of ancestral karyotype blocks in the Sinapis alba L. genome. Genome 54:33–41
Panjabi P, Jagannath A, Bisht NC, Padmaja KL, Sharma S, Gupta V, Pradhan AK, Pental D (2008) Comparative mapping of Brassica juncea and Arabidopsis thaliana using intron polymorphism (IP) markers: homoeologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genom 9:113
Quraishi UM, Abrouk M, Bolot S, Pont C, Throude M, Guilhot N, Confolent C, Bortolini F, Praud S, Murigneux A, Charmet G, Salse J (2009) Genomics in cereals: from genome-wide conserved orthologous set (COS) sequences to candidate genes for trait dissection. Funct Integr Genomics 9:473–484
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Schranz ME, Lysak MA, Mitchell-Olds T (2006) The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci 11:535–542
Shirasawa K, Oyama M, Hirakawa H, Sato S, Tabata S, Fujioka T, Kimizuka-Takagi C, Sasamoto S, Watanabe A, Kato M, Kishida Y, Kohara M, Takahashi C, Tsuruoka H, Wada T, Sakai T, Isobe S (2011) An EST-SSR linkage map of Raphanus sativus and comparative genomics of the Brassicaceae. DNA Res 18:221–232
Slotte T, Hazzouri K, Ågren J, Koenig D, Maumus F, Guo Y, Steige K, Platts A, Escobar J, Newman L, Wang W, Mandáková T, Vello E, Smith L, Henz S, Steffen J, Takuno S, Brandvain Y, Coop G, Andolfatto P, Hu T, Blanchette M, Clark R, Quesneville H, Nordborg M, Gaut B, Lysak M, Jenkins J, Grimwood J, Chapman J, Prochnik S, Shu S, Rokhsar D, Schmutz J, Weigel D, Wright S (2013) The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat Genet 45:831–835
Song KM, Osborn TC, Williams PH (1988) Brassica taxonomy based on nuclear restriction fragment length polymorphisms (RFLPs): 2. Preliminary analysis of subspecies within B. rapa (syn. campestris) and B. oleracea. Theor Appl Genet 76:593–600
Song K, Osborn TC, Williams PH (1990) Brassica taxonomy based on nuclear restriction fragment length polymorphisms (RFLPs): 3. Genome relationships in Brassica and related genera and the origin of B. oleracea and B. rapa (syn. campestris). Theor Appl Genet 79:497–506
The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
The Brassica rapa Genome Sequencing Project Consortium (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43:1035–1039
Thormann CE, Ferreira ME, Camargo LE, Tivang JG, Osborn TC (1994) Comparison of RFLP and RAPD markers to estimating genetic relationships within and among cruciferous species. Theor Appl Genet 88:973–980
Timms L, Jimenez R, Chase M, Lavelle D, McHale L, Kozik A, Lai Z, Heesacker A, Knapp S, Rieseberg L, Michelmore R, Kesseli R (2006) Analyses of synteny between Arabidopsis thaliana and species in the Asteraceae reveal a complex network of small syntenic segments and major chromosomal rearrangements. Genetics 173:2227–2235
van Ooijen JW (2006) JoinMap® 4, software for the calculation of genetic linkage maps in experimental populations. Kyazma B. V, Wageningen
Warwick S, Black L (1991) Molecular systematics of Brassica and allied genera (Subtribe Brassicinae, Brassiceae)—chloroplast genome and cytodeme congruence. Theor Appl Genet 82:81–92
Warwick S, Black L (1997) Phylogenetic implications of chloroplast DNA restriction site variation in subtribes Raphaninae and Cakilinae (Brassicaceae, tribe Brassiceae). Canadian J Bot 75:960–973
Wu F, Mueller LA, Crouzillat D, Petiard V, Tanksley SD (2006) Combining bioinformatics and phylogenetics to identify large sets of single-copy orthologous genes (COSII) for comparative, evolutionary and systematic studies: a test case in the euasterid plant clade. Genetics 174:1407–1420
Wu F, Eannetta NT, Xu Y, Durrett R, Mazourek M, Jahn MM, Tanksley SD (2009a) A COSII genetic map of the pepper genome provides a detailed picture of synteny with tomato and new insights into recent chromosome evolution in the genus Capsicum. Theor Appl Genet 118:1279–1293
Wu F, Eannetta NT, Xu Y, Tanksley SD (2009b) A detailed synteny map of the eggplant genome based on conserved ortholog set II (COSII) markers. Theor Appl Genet 118:927–935
Wu F, Eannetta NT, Xu Y, Plieske J, Ganal M, Pozzi C, Bakaher N, Tanksley SD (2010) COSII genetic maps of two diploid Nicotiana species provide a detailed picture of synteny with tomato and insights into chromosome evolution in tetraploid N. tabacum. Theor Appl Genet 120:809–827
Wu HJ, Zhang Z, Wang JY, Oh DH, Dassanayake M, Liu B, Huang Q, Sun HX, Xia R, Wu Y, Wang YN, Yang Z, Liu Y, Zhang W, Zhang H, Chu J, Yan C, Fang S, Zhang J, Wang Y, Zhang F, Wang G, Lee SY, Cheeseman JM, Yang B, Li B, Min J, Yang L, Wang J, Chu C, Chen SY, Bohnert HJ, Zhu JK, Wang XJ, Xie Q (2012) Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc Natl Acad Sci USA 109:12219–12224
Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Yang YW, Lai KN, Tai PY, Ma DP, Li WH (1999) Molecular phylogenetic studies of Brassica, Rrippa, Arabidopsis and allied genera based on the internal transcribed spacer region of 18S–25S rDNA. Mol Phylogenet Evol 13:455–462
Yang YW, Tai PY, Chen Y, Li WH (2002) A study of the phylogeny of Brassica rapa, B. nigra, Raphanus sativus, and their related genera using noncoding regions of chloroplast DNA. Mol Phylogene Evol 23:268–275
Yang R, Jarvis DE, Chen H, Beilstein MA, Grimwood J, Jenkins J, Shu S, Prochnik S, Xin M, Ma C, Schmutz J, Wing RA, Mitchell-Olds T, Schumaker KS, Wang X (2013) The reference genome of the halophytic plant Eutrema salsugineum. Front Plant Sci 4:46
Acknowledgments
This work was supported by grants from the Next-Generation Biogreen21 program (PJ008019), Rural Development Administration, Korea to HJY and 2013 Research Fund of Myongji University to JHM. We thank Dr. Shengyi Liu (Oil Crops Research Institute of CAAS, China) for kindly providing sequence information of Brassica oleracea, Sin-Gi Park (National Academy of Agricultural Science of RDA, Korea) for bioinformatics support, and Dr. Suhyoung Park (National Institute of Horticultural and Herbal Science of RDA, Korea) for providing plant material.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The authors declare that the experiments complied with current laws of the country in which they were performed.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by Isobel Parkin.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. S1 Functional classification of the Brassica COS genes based on gene ontology mapping using TAIR database. GO of whole At genes was compared as control and Z test was used to examine statistical analysis. A. Cellular component categories. B. Molecular function categories. C. Biological process categories.
Supplemental Fig. S2 PCR amplification of six COS genes in selected species of Brassicaceae. At, A. thaliana; Rs, R. sativus; A, B. rapa (A genome); B, B. nigra (B genome); C, B. olearacea (C genome); AB, B. juncea (AB genome); AC, B. napus (AC genome); BC, B. carinata (BC genome); A + B, genomic DNA mixture of A and B; A + C, genomic DNA mixture of A and C; B + C, genomic DNA mixture of B and C.
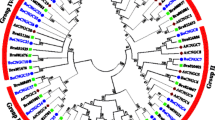
Supplemental Fig. S3 Phylogenetic tree of 13 species of Brassicaceae. Tree was generated using MEGA5 program by Maximum Likelihood analysis of COS0015, COS245, and COS566 genes. The stability of tree nodes was tested by bootstrap analysis with 1,000 replicates. Bootstrap values are indicated on the branches and the branch length reflects the estimated number of substitutions per 100 sites. A local cluster of 5 wild Mediterranean Brassica species, Br, and Bo is indicated in a green box.
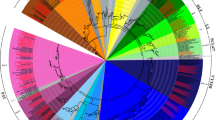
Supplemental Fig. S4 Circos diagram of syntenic genome block pairs between Rs and Br. Conserved orthologous blocks in Rs are plotted against their syntenic counterpart in Br. The numbers in Rs linkage groups indicate genetic distance in cM whereas those in Br chromosomes indicate 5 Mb intervals. The syntenic counterparts of conserved blocks between the genomes are interconnected by colored lines. Br, B. rapa; Rs, R. sativus.
Supplemental Fig. S5 Circos diagram of COS gene pairs between Rs and Br corresponding eight ancient chromosomes. A. AK1, B. AK2, C. AK3, D. AK4, E. AK5, F. AK6, G. AK7, H. AK8. Conserved orthologous genes in Rs are plotted against their syntenic counterpart in Br. The numbers in Rs linkage groups indicate genetic distance in cM whereas those in Br chromosomes indicate 5 Mb intervals. The syntenic counterparts of conserved gene pair between the genomes are interconnected by colored lines. The line colors indicate three differentially fractionated subgenome types in the Br genome. Br, B. rapa; Rs, R. sativus.
Rights and permissions
About this article
Cite this article
Jeong, YM., Chung, WH., Chung, H. et al. Comparative analysis of the radish genome based on a conserved ortholog set (COS) of Brassica . Theor Appl Genet 127, 1975–1989 (2014). https://doi.org/10.1007/s00122-014-2354-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-014-2354-3