Abstract
Key message
Considerable genome variation had been incorporated within rapeseed breeding programs over past decades.
Abstract
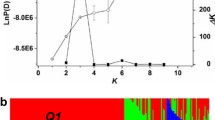
In past decades, there have been substantial changes in phenotypic properties of rapeseed as a result of extensive breeding effort. Uncovering the underlying patterns of allelic variation in the context of genome organisation would provide knowledge to guide future genetic improvement. We assessed genome-wide genetic changes, including population structure, genetic relatedness, the extent of linkage disequilibrium, nucleotide diversity and genetic differentiation based on F ST outlier detection, for a panel of 472 Brassica napus inbred accessions using a 60 k Brassica Infinium® SNP array. We found genetic diversity varied in different sub-groups. Moreover, the genetic diversity increased from 1950 to 1980 and then remained at a similar level in China and Europe. We also found ~6–10 % genomic regions revealed high F ST values. Some QTLs previously associated with important agronomic traits overlapped with these regions. Overall, the B. napus C genome was found to have more high F ST signals than the A genome, and we concluded that the C genome may contribute more valuable alleles to generate elite traits. The results of this study indicate that considerable genome variation had been incorporated within rapeseed breeding programs over past decades. These results also contribute to understanding the impact of rapeseed improvement on available genome variation and the potential for dissecting complex agronomic traits.







Similar content being viewed by others
References
Abbadi A, Leckband G (2011) Rapeseed breeding for oil content, quality, and sustainability. Eur J Lipid Sci Technol 113:1198–1206
Albrechtsen A, Nielsen FC, Nielsen R (2010) Ascertainment biases in SNP chips affect measures of population divergence. Mol Biol Evol 27:2534–2547
Allender CJ, King GJ (2010) Origins of the amphiploid species Brassica napus L. investigated by chloroplast and nuclear molecular markers. BMC Plant Biol 10:54
Bancroft I, Morgan C, Fraser F, Higgins J, Wells R, Clissold L, Baker D, Long Y, Meng J, Wang X, Liu S, Trick M (2011) Dissecting the genome of the polyploid crop oilseed rape by transcriptome sequencing. Nat Biotechnol 29:762–766
Becker HC, Engqvist GM, Karlsson B (1995) Comparison of rapeseed cultivars and resynthesized lines based on allozyme and RFLP markers. Theor Appl Genet 91:62–67
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Bus A, Korber N, Snowdon RJ, Stich B (2011) Patterns of molecular variation in a species-wide germplasm set of Brassica napus. Theor Appl Genet 123:1413–1423
Cai D, Xiao Y, Yang W, Ye W, Wang B, Younas M, Wu J, Liu K (2014) Association mapping of six yield-related traits in rapeseed (Brassica napus L.). Theor Appl Genet 127:85–96
Cao Z, Tian F, Wang N, Jiang C, Lin B, Xia W, Shi J, Long Y, Zhang C, Meng J (2010) Analysis of QTLs for erucic acid and oil content in seeds on A8 chromosome and the linkage drag between the alleles for the two traits in Brassica napus. J Genet Genomics 37:231–240
Cavanagh CR, Chao S, Wang S, Huang BE, Stephen S, Kiani S, Forrest K, Saintenac C, Brown-Guedira GL, Akhunova A, See D, Bai G, Pumphrey M, Tomar L, Wong D, Kong S, Reynolds M, da Silva ML, Bockelman H, Talbert L, Anderson JA, Dreisigacker S, Baenziger S, Carter A, Korzun V, Morrell PL, Dubcovsky J, Morell MK, Sorrells ME, Hayden MJ, Akhunov E (2013) Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc Natl Acad Sci USA 110:8057–8062
Chen ZJ, Pikaard CS (1997) Transcriptional analysis of nucleolar dominance in polyploid plants: biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. Proc Natl Acad Sci USA 94:3442–3447
Chen S, Nelson MN, Ghamkhar K, Fu T, Cowling WA (2008) Divergent patterns of allelic diversity from similar origins: the case of oilseed rape (Brassica napus L.) in China and Australia. Genome 51:1–10
Delourme R, Chèvre AM, Brun H, Rouxel T, Balesdent MH, Dias JS, Salisbury P, Renard M, Rimmer SR (2006) Major gene and polygenic resistance to leptosphaeria maculans in oilseed rape (Brassica napus). Eur J Plant Pathol 114:41–52
Duran C, Eales D, Marshall D, Imelfort M, Stiller J, Berkman PJ, Clark T, McKenzie M, Appleby N, Batley J, Basford K, Edwards D (2010) Future tools for association mapping in crop plants. Genome 53:1017–1023
Ecke W, Clemens R, Honsdorf N, Becker HC (2010) Extent and structure of linkage disequilibrium in canola quality winter rapeseed (Brassica napus L.). Theor Appl Genet 120:921–931
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Fu Y-B, Somers DJ (2009) Genome-wide reduction of genetic diversity in wheat breeding. Crop Sci 49:161–168
Georges M (1997) QTL mapping to QTL cloning: mice to the rescue. Genome Res 7:663–665
Gore MA, Chia JM, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL, Peiffer JA, McMullen MD, Grills GS, Ross-Ibarra J, Ware DH, Buckler ES (2009) A first-generation haplotype map of maize. Science 326:1115–1117
Guo B, Wang D, Guo Z, Beavis WD (2013) Family-based association mapping in crop species. Theor Appl Genet 126:1419–1430
Harper AL, Trick M, Higgins J, Fraser F, Clissold L, Wells R, Hattori C, Werner P, Bancroft I (2012) Associative transcriptomics of traits in the polyploid crop species Brassica napus. Nat Biotechnol 30:798–802
Hasan M, Friedt W, Pons-Kuhnemann J, Freitag NM, Link K, Snowdon RJ (2008) Association of gene-linked SSR markers to seed glucosinolate content in oilseed rape (Brassica napus). Theor Appl Genet 116:1035–1049
Hyten DL, Song Q, Zhu Y, Choi IY, Nelson RL, Costa JM, Specht JE, Shoemaker RC, Cregan PB (2006) Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci USA 103:16666–16671
Jiao Y, Zhao H, Ren L, Song W, Zeng B, Guo J, Wang B, Liu Z, Chen J, Li W, Zhang M, Xie S, Lai J (2012) Genome-wide genetic changes during modern breeding of maize. Nat Genet 44:812–815
Kim S, Plagnol V, Hu TT, Toomajian C, Clark RM, Ossowski S, Ecker JR, Weigel D, Nordborg M (2007) Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet 39:1151–1155
Lai J, Li R, Xu X, Jin W, Xu M, Zhao H, Xiang Z, Song W, Ying K, Zhang M, Jiao Y, Ni P, Zhang J, Li D, Guo X, Ye K, Jian M, Wang B, Zheng H, Liang H, Zhang X, Wang S, Chen S, Li J, Fu Y, Springer NM, Yang H, Wang J, Dai J, Schnable PS, Wang J (2010) Genome-wide patterns of genetic variation among elite maize inbred lines. Nat Genet 42:1027–1030
Lam HM, Xu X, Liu X, Chen W, Yang G, Wong FL, Li MW, He W, Qin N, Wang B, Li J, Jian M, Wang J, Shao G, Wang J, Sun SS, Zhang G (2010) Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet 42:1053–1059
Lefebvre DD (1989) Increased potassium absorption confers resistance to group IA cations in rubidium-selected suspension cells of Brassica napus. Plant Physiol 91:1460–1466
Li M, Liu J, Wang Y, Yu L, Meng J (2007) Production of partial new-typed Brassica napus by introgression of genomic components from B. rapa and B. carinata. J Genet Genomics 34:460–468
Li F, Chen B, Xu K, Wu J, Song W, Bancroft I, Harper AL, Trick M, Liu S, Gao G, Wang N, Yan G, Qiao J, Li J, Li H, Xiao X, Zhang T, Wu X (2014) Genome-wide association study dissects the genetic architecture of seed weight and seed quality in rapeseed (Brassica napus L.). DNA Res. doi:10.1093/dnares/dsu002
Lu HY, Liu XF, Wei SP, Zhang YM (2011) Epistatic association mapping in homozygous crop cultivars. PLoS One 6:e17773
McCouch SR, Doerge RW (1995) QTL mapping in rice. Trends Genet 11:482–487
McNally KL, Childs KL, Bohnert R, Davidson RM, Zhao K, Ulat VJ, Zeller G, Clark RM, Hoen DR, Bureau TE, Stokowski R, Ballinger DG, Frazer KA, Cox DR, Padhukasahasram B, Bustamante CD, Weigel D, Mackill DJ, Bruskiewich RM, Ratsch G, Buell CR, Leung H, Leach JE (2009) Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc Natl Acad Sci USA 106:12273–12278
Mei J, Li Q, Qian L, Fu Y, Li J, Frauen M, Qian W (2011) Genetic investigation of the origination of allopolyploid with virtually synthesized lines: application to the C subgenome of Brassica napus. Heredity 106:955–961
Morrell PL, Buckler ES, Ross-Ibarra J (2011) Crop genomics: advances and applications. Nat Rev Genet 13:85–96
Motsinger AA, Reif DM, Dudek SM, Ritchie MD (2006) Understanding the evolutionary process of grammatical evolution neural networks for feature selection in genetic epidemiology. Proc IEEE Symp Comput Intell Bioinform Comput Biol 2006:1–8
Nei M, Miller JC (1990) A simple method for estimating average number of nucleotide substitutions within and between populations from restriction data. Genetics 125:873–879
Nicholson G, Smith AV, Jónsson F, Gstafsson Ã, Stefánsson K, Donnelly P (2002) Assessing population differentiation and isolation from single-nucleotide polymorphism data. J R Stat Soc 64:695–715
Porto-Neto LR, Lee SH, Lee HK, Gondro C (2013) Detection of signatures of selection using FST. Methods Mol Biol 1019:423–436
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575
Qian W, Meng J, Li M, Frauen M, Sass O, Noack J, Jung C (2006) Introgression of genomic components from Chinese Brassica rapa contributes to widening the genetic diversity in rapeseed (B. napus L.), with emphasis on the evolution of Chinese rapeseed. Theor Appl Genet 113:49–54
Richards E, Reichardt M, Rogers S (2001) Preparation of genomic DNA from plant tissue. Curr Protoc Mol Biol, chapter 2, unit 2.3
Rygulla W, Snowdon RJ, Eynck C, Koopmann B, von Tiedemann A, Luhs W, Friedt W (2007) Broadening the genetic basis of Verticillium longisporum resistance in Brassica napus by interspecific hybridization. Phytopathology 97:1391–1396
Shahidi F (1990) Canola and rapeseed: production, chemistry, nutrition and processing technology. Van Nostrand Reinhold, New York, pp 1–10
Shi J, Li R, Qiu D, Jiang C, Long Y, Morgan C, Bancroft I, Zhao J, Meng J (2009) Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus. Genetics 182:851–861
Sim SC, Van Deynze A, Stoffel K, Douches DS, Zarka D, Ganal MW, Chetelat RT, Hutton SF, Scott JW, Gardner RG, Panthee DR, Mutschler M, Myers JR, Francis DM (2012) High-density SNP genotyping of tomato (Solanum lycopersicum L.) reveals patterns of genetic variation due to breeding. PLoS One 7:e45520
Song K, Osborn TC (1992) Polyphyletic origins of Brassica napus: new evidence based on organelle and nuclear RFLP analyses. Genome 35:992–1001
Stefansson BR, Hougen FW (1964) Selection of rape plants (Brassica napus) with seed oil practically free from erucic acid. Can J Plant Sci 44:359–364
Suzuki Y (2010) Statistical methods for detecting natural selection from genomic data. Genes Genet Syst 85:359–376
U N (1935) Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn J Bot 7:389–452
Vaz Patto MC, Rubiales D, Martin A, Hernandez P, Lindhout P, Niks RE, Stam P (2003) QTL mapping provides evidence for lack of association of the avoidance of leaf rust in Hordeum chilense with stomata density. Theor Appl Genet 106:1283–1292
Wang J, Chen Z, Du J, Sun Y, Liang A (2005) Novel insect resistance in Brassica napus developed by transformation of chitinase and scorpion toxin genes. Plant Cell Rep 24:549–555
Wang N, Wang Y, Tian F, King GJ, Zhang C, Long Y, Shi L, Meng J (2008) A functional genomics resource for Brassica napus: development of an EMS mutagenized population and discovery of FAE1 point mutations by TILLING. New Phytol 180:751–765
Wang J, Long Y, Wu B, Liu J, Jiang C, Shi L, Zhao J, King GJ, Meng J (2009) The evolution of Brassica napus FLOWERING LOCUS T paralogues in the context of inverted chromosomal duplication blocks. BMC Evol Biol 9:271
Wang N, Shi L, Tian F, Ning H, Wu X, Long Y, Meng J (2010) Assessment of FAE1 polymorphisms in three Brassica species using EcoTILLING and their association with differences in seed erucic acid contents. BMC Plant Biol 10:137
Waugh R, Marshall D, Thomas B, Comadran J, Russell J, Close T, Stein N, Hayes P, Muehlbauer G, Cockram J, O’Sullivan D, Mackay I, Flavell A, Agoueb, BarleyCap, Ramsay L (2010) Whole-genome association mapping in elite inbred crop varieties. Genome 53:967–972
Wu J, Cai G, Tu J, Li L, Liu S, Luo X, Zhou L, Fan C, Zhou Y (2013) Identification of QTLs for resistance to sclerotinia stem rot and BnaC.IGMT5.a as a candidate gene of the major resistant QTL SRC6 in Brassica napus. PLoS One 8:e67740
Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88:76–82
Zhu Q, Zheng X, Luo J, Gaut BS, Ge S (2007) Multilocus analysis of nucleotide variation of Oryza sativa and its wild relatives: severe bottleneck during domestication of rice. Mol Biol Evol 24:875–888
Acknowledgments
This work was supported by the Chinese National Basic Research and Development Program (2011CB109302), National Science & Technology Pillar Program during the Twelfth Five-year Plan Period (2013BAD01B03 and 2011BAD35B09) and Crop Germplasm Protection Project (NB2011-2130135). GK is supported in part by the Chutian Scholar program of Hubei province.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
The authors declare that the study comply with the current laws of the country in which they were performed.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Michael Gore.
N. Wang and F. Li have contributed equally.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2014_2343_MOESM4_ESM.tif
Genomic distribution of putative artificial selection among sub-populations classified by five breeding regions. Signatures of putative artificial selection were measured by FST based on calculation of genetic differentiation among sub-populations five from five breeding regions. From the top to bottom, these five panels are corresponding to sub-populations of Australia, China, Europe, North America and Northeast Asia, respectively. FST was smoothed with a 0.5 Mb window and a 50 kb siding bin (see M&M). The red dash lines indicate value of 1st percentile of the empirical distribution of high FST-windows. Chromosomes were differed by black and gray plots sequentially (TIFF 1588 kb)
122_2014_2343_MOESM5_ESM.tif
Genomic distribution of putative artificial selection among sub-populations classified by three growth types. From the top to bottom, these three panels are corresponding to sub-populations of spring, semi-winter and winter, respectively. All other information is similar with Figure S4 (TIFF 1226 kb)
122_2014_2343_MOESM6_ESM.tif
Genomic distribution of putative artificial selection among sub-populations classified by five breeding periods in China. From the top to the bottom, these five panels are corresponding to sub-populations of 1950-1970, 1971-1980, 1981-1990, 1991-2000 and 2001-2011, respectively. All other information is similar with Figure S4. (TIFF 1723 kb)
122_2014_2343_MOESM7_ESM.tif
Genomic distribution of putative artificial selection among sub-populations classified by four breeding period in Europe. From the top to the bottom, these four panels are corresponding to sub-populations of 1950-1970, 1971-1980, 1981-1990 and 1991-2000, respectively. All other information is similar with Figure S4. (TIFF 1260 kb)
122_2014_2343_MOESM13_ESM.xlsx
Genomic regions with outlier FST windows based on sub-populations classified by different breeding periods in China and Europe (XLSX 17 kb)
Rights and permissions
About this article
Cite this article
Wang, N., Li, F., Chen, B. et al. Genome-wide investigation of genetic changes during modern breeding of Brassica napus . Theor Appl Genet 127, 1817–1829 (2014). https://doi.org/10.1007/s00122-014-2343-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-014-2343-6