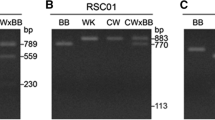
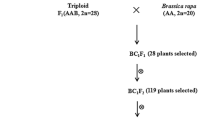
Abstract
A hybridization barrier leads to the inability of seed formation after intergeneric crossings between Brassica rapa and Raphanus sativus. Most B. rapa lines cannot set intergeneric hybrid seeds because of embryo breakdown, but a B. rapa line obtained from turnip cultivar ‘Shogoin-kabu’ is able to produce a large number of hybrid seeds as a maternal parent by crossings with R. sativus. In ‘Shogoin-kabu’ crossed with R. sativus, developments of embryos and endosperms were slower than those in intraspecific crossings, but some of them grew to mature seeds without embryo breakdown. Intergeneric hybrid seeds were obtained in a ‘Shogoin-kabu’ line at a rate of 0.13 per pollinated flower, while no hybrid seeds were obtained in a line developed from Chinese cabbage cultivar ‘Chiifu’. F1 hybrid plants between the lines of ‘Shogoin-kabu’ and ‘Chiifu’ set a larger number of hybrid seeds per flower, 0.68, than both the parental lines. Quantitative trait loci (QTLs) for hybrid seed formation were analyzed after intergeneric crossings using two different F2 populations derived from the F1 hybrids, and three QTLs with significant logarithm of odds scores were detected. Among them, two QTLs, i.e., one in linkage group A10 and the other in linkage group A01, were detected in both the F2 populations. These two QTLs had contrary effects on the number of hybrid seeds. Epistatic interaction between these two QTLs was revealed. Possible candidate genes controlling hybrid seed formation ability in QTL regions were inferred using the published B. rapa genome sequences.






Similar content being viewed by others
References
Akaba M, Kaneko Y, Hatakeyama K, Ishida M, Bang SW, Matsuzawa Y (2009) Identification and evaluation of clubroot resistance of radish chromosome using a Brassica napus-Raphanus sativus monosomic addition line. Breed Sci 59:203–206
Berger F, Chaudhury A (2009) Parental memories shape seeds. Trends Plant Sci 14:550–556
Brown J, Brown AP, Davis JB, Erickson D (1997) Intergeneric hybridization between Sinapis alba and Brassica napus. Euphytica 93:163–168
Bushell C, Spielman M, Scott RJ (2003) The basis of natural and artificial postzygotic hybridization barriers in Arabidopsis species. Plant Cell 15:1430–1442
de Nettancourt D (2001) Incompatibility and incongruity in wild and cultivated plants, 2nd edn. Springer, Berlin, p 322
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Dresselhaus T, Márton ML (2009) Micropylar pollen tube guidance and burst: adapted from defense mechanisms? Curr Opin Plant Biol 12:773–780
Evans MMS, Kermicle JL (2001) Teosinte crossing barrier1, a locus governing hybridization of teosinte with maize. Theor Appl Genet 103:259–265
Gehring M, Bubb KL, Henikoff S (2009) Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324:1447–1451
Guitton AE, Page DR, Chambrier P, Lionnet C, Faure JE, Grossniklaus U, Berger F (2004) Identification of new members of Fertilization Independent Seed Polycomb Group pathway involved in the control of seed development in Arabidopsis thaliana. Development 131:2971–2981
Hennig L, Bouveret R, Gruissem W (2005) MSI1-like proteins: an escort service for chromatin assembly and remodeling complexes. Trends Cell Biol 15:295–302
Holec S, Berger F (2012) Polycomb group complexes mediate developmental transitions in plants. Plant Physiol 158:35–43
Hsieh TF, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D (2009) Genome-wide demethylation of Arabidopsis endosperm. Science 324:1451–1454
Huh JH, Bauer MJ, Hsieh TF, Fischer RL (2008) Cellular programming of plant gene imprinting. Cell 132:735–744
Johnston SA, Hanneman REJ (1982) Manipulations of endosperm balance number overcome crossing barriers between diploid Solanum species. Science 217:446–448
Johnston SA, Nijs TPM, Peloquin SJ, Hanneman RE (1980) The significance of genic balance to endosperm development in interspecific crosses. Theor Appl Genet 57:5–9
Jossefson C, Dikes B, Comai L (2006) Parent-dependent loss of gene silencing during interspecies hybridization. Curr Biol 16:1322–1328
Jullien PE, Berger F (2010) Parental genome dosage imbalance deregulates imprinting in Arabidopsis. PLoS Genet 6:e1000885
Kaneko Y, Matsuzawa Y, Namai H, Sarashima M (1993) Genetical and breeding evaluation of chromosome addition lines of radish with single kale chromosome. I. Phenotypic expression of some monosomic addition lines for radish and turnip varieties. Bull Coll Agric Utsunomiya Univ 15:27–37
Kang IH, Steffen JG, Portereiko MF, Lloyd A, Drews GN (2008) The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20:635–647
Karpechenko GD (1924) Hybrids of Raphanus sativus L. × Brassica oleracea L. J Genet 14:375–396
Kermicle JL, Evans MMS (2010) The Zea mays sexual compatibility gene ga2: Naturally occurring alleles, their distribution, and role in reproductive isolation. J Heredity 101:737–749
Kinoshita T (2007) Reproductive barrier and genomic imprinting in the endosperm of flowering plants. Genes Genet Syst 82:177–186
Köhler C, Hennig L, Bouveret R, Gheyselinck J, Grossniklaus U, Gruissem W (2003) Arabidopsis MSI1 is a component of the MEA/FIE polycomb group complex and required for seed development. EMBO J 22:4804–4814
Koizuka N, Imai R, Fujimoto H, Hayakawa T, Kimura Y, Kohno-Murase J, Sakai T, Kawasaki S, Imamura J (2003) Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restore fertility in the cytoplasmic male-sterile Kosena redish. Plant J 34:407–415
Li F, Kitashiba H, Inaba K, Nishio T (2009) A Brassica rapa linkage map of EST-based SNP markers for identification of candidate genes controlling flowering time and leaf morphological traits. DNA Res 16:311–323
Li F, Hasegawa Y, Saito M, Shirasawa S, Fukushima A, Ito T, Fujii H, Kishitani S, Kitashiba H, Nishio T (2011) Extensive chromosome homoeology among Brassiceae species were revealed by comparative genetic mapping with high-density EST-based SNP markers in radish (Raphanus sativus L.). DNA Res 18:401–411
Lowry DB, Modliszeqski L, Wright KM, Wu CA, Willis JH (2008) The strength and genetic basis of reproductive isolating barrier in flowering plants. Phil Trans R Soc B 363:3009–3022
Nishiyama I, Inomata N (1966) Embryological studies on cross-incompatibility between 2x and 4x in Brassica. Japan J Genet 41:27–42
Nishiyama I, Yabuno T (1978) Causal relationships between the polar nuclei in double fertilization and interspecific cross-incompatibility in Avena. Cytologia 43:453–466
Scott RJ, Spielman M, Bailey J, Dickinson HG (1998) Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125:3329–3341
Sharma HC, Gill BS (1983) Current status of wide hybridization in wheat. Euphytica 32:17–31
Shiokai S, Shirasawa K, Sato Y, Nishio T (2010) Improvement of the dot-blot-SNP technique for efficient and cost-effective genotyping. Mol Breed 25:179–185
Shirasawa K, Shiokai S, Yamaguchi M, Kishitani S, Nishio T (2006) Dot-blot-SNP analysis for practical plant breeding and cultivar identification in rice. Theor Appl Genet 113:147–155
Stoute A, Varenko V, King GJ, Scott RJ, Kurup S (2012) Parental genome imbalance in Brassica oleracea causes asymmetric triploid block. Plant J 71:503–516
Takahata Y, Takeda T (1990) Intergeneric (intersubtribe) hybridization between Moricandia arvensis and Brassica A and B genome species by ovary culture. Theor Appl Genet 80:38–42
Tonosaki K, Nishio T (2010) Identification of species in tribe Brassiceae by dot-blot hybridization using species-specific ITS1 probes. Plant Cell Rep 29:1179–1186
Udagawa H, Ishimaru Y, Li F, Sato Y, Kitashiba H, Nishio T (2010) Genetic analysis of interspecific incompatibility in Brassica rapa. Theor Appl Genet 121:689–696
Van Ooijen JW (2006) JoinMap ver.4, Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen
Wang YP, Luo P (1998) Intergeneric hybridization between Brassica species and Crambe abyssinica. Euphytica 101:1–7
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S et al (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43:1035–1039
Acknowledgments
We are grateful to Dr. Ashtosh for careful reading of a manuscript. This work was supported in part by the Program for Promotion of Basic and Applied Research for Innovations in Bio-oriented Industry (BRAIN), Japan and JSPS KAKENHI Grand Number 24380002.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by I. Paran.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2012_2021_MOESM3_ESM.pdf
Supplementary material 3 (PDF 92 kb) Leaves (a–d) and flowers (e–f) of parents and intergeneric hybrids. A, E: B. rapa ‘Chiifu’. B, F: B. rapa ‘Shogoin-kabu’. C, G: R. sativus ‘Shogoin-daikon’. D, H: intergeneric hybrid
122_2012_2021_MOESM4_ESM.pdf
Supplementary material 4 (PDF 27 kb)Distribution of the numbers of hybrid seeds per pollinated flower in F2 populations used in 2008 (a) and 2010 (b). Black arrows indicate the ‘Chiifu’ line, arrowheads indicate the ‘Shogoin-kabu’ line, and white arrows represent F1 plants between the ‘Shogoin-kabu’ and ‘Chiifu’ lines
Rights and permissions
About this article
Cite this article
Tonosaki, K., Michiba, K., Bang, S.W. et al. Genetic analysis of hybrid seed formation ability of Brassica rapa in intergeneric crossings with Raphanus sativus . Theor Appl Genet 126, 837–846 (2013). https://doi.org/10.1007/s00122-012-2021-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-012-2021-5