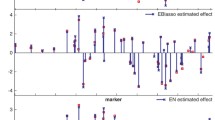
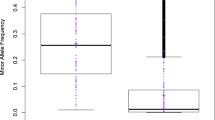
Abstract
Introduction
Virtually all existing expectation-maximization (EM) algorithms for quantitative trait locus (QTL) mapping overlook the covariance structure of genetic effects, even though this information can help enhance the robustness of model-based inferences.
Results
Here, we propose fast EM and pseudo-EM-based procedures for Bayesian shrinkage analysis of QTLs, designed to accommodate the posterior covariance structure of genetic effects through a block-updating scheme. That is, updating all genetic effects simultaneously through many cycles of iterations.
Conclusion
Simulation results based on computer-generated and real-world marker data demonstrated the ability of our method to swiftly produce sensible results regarding the phenotype-to-genotype association. Our new method provides a robust and remarkably fast alternative to full Bayesian estimation in high-dimensional models where the computational burden associated with Markov chain Monte Carlo simulation is often unwieldy. The R code used to fit the model to the data is provided in the online supplementary material.




Similar content being viewed by others
References
Ball RD (2001) Bayesian methods for quantitative trait loci mapping based on model selection: approximate analysis using Bayesian information criterion. Genetics 159:1351–1364
Bishop CM, Tipping ME (2003) Bayesian regression and classification. In: Suykens J, Horvath G, Basu S, Micchelli C, Vandewalle J (eds) Advances in learning theory: methods, models and applications, vol 190. IOS Press, NATO Science, Amsterdam, pp 267–285
Broman KW, Speed TP (2002) A model selection approach for the identification of quantitative trait loci in experimental crosses (with discussion). J Roy Stat Soc B 64:641–656
Broman KW (2001) Review of statistical methods for QTL mapping in experimental crosses. Lab Anim 30:44–52
Cai X, Huang A, Xu S (2011) Fast empirical Bayesian LASSO for multiple quantitative trait locus mapping. BMC Bioinform 12:211
Carbonell EA, Asins MJ, Baselga M, Balansard E, Gerig TM (1993) Power studies in the estimation of genetic parameters and the localization of quantitative trait loci for backcross and doubled haploid populations. Theor Appl Genet 86:411–416
Carlborg Ö, Andersson L (2002) Use of randomization testing to detect multiple epistatic QTLs. Genet Sel Evol 79:175–184
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Cleveland MA, Forni S, Nader D, Maltecca C (2010) Genomic breeding value prediction using three Bayesian methods and application to reduced density marker panels. BMC Proc 4(Suppl 1):S6
Conti DV, Witte J (2003) Hierarchical modeling of linkage disequilibrium: genetic structure and spatial relations. Am J Hum Genet 72:351–363
de los Campos G, Naya H, Gianola D, Crossa J, Legarra A, Manfredi E, Weigel K, Cotes JM (2009) Predicting quantitative traits with regression models for dense molecular markers and pedigree. Genetics 182:375–385
Dempster AP, Laird NM, Rubin DB (1977) Maximum likelihood from incomplete data via the EM algorithm. J Roy Stat Soc B 39:1–38
Fridley BL, Jenkins GD (2010) Localizing putative markers in genetic association studies by incorporating linkage disequilibrium into Bayesian hierarchical models. Hum Hered 70:63–73
Gelman A, Hill J (2007) Data analysis using regression and multilevel/hierarchical models. Cambridge University Press, New York
Gelman A, Carlin JB, Stern HS, Rubin DB (2003) Bayesian data analysis, 2nd edn. Chapman and Hall, New York
George EI, McCulloch RE (1993) Variable selection via Gibbs sampling. J Am Stat Assoc 88:881–889
Gilks WR, Richardson S, Spiegelhalter DJ (eds) (1996) Markov Chain Monte Carlo in practice. Chapman and Hall, London
Gimelfarb A, Lande R (1994a) Simulation of marker-assisted selection in hybrid populations. Genet Res 63:39–47
Gimelfarb A, Lande R (1994b) Simulation of marker-assisted selection for non-additive traits. Genet Res 64:127–136
Golub G, van Loan C (1996) Matrix computations, 3rd edn. The John Hopkins University Press, Baltimore
Hayashi T, Iwata H (2010) EM algorithm for Bayesian estimation of genomic breeding values. BMC Genet 11:3
Heckerman D, Chickering DM, Meek C, Rounthwaite R, Kadie C (2000) Dependency network for inference, collaborative filtering, and data visualization. J Mach Learn Res 1:49–75
Henderson CR (1950) Estimation of genetic parameters. Ann Math Stat 21:309–310
Henderson CR (1970) Best linear unbiased estimation and prediction under a selection model. Biometrics 31:423–447
Hoerl AE, Kennard RW (1970) Ridge regression: biased estimation for nonorthogonal problems. Technometrics 12:55–67
Hoti F, Sillanpää MJ (2006) Bayesian mapping of genotype × expression interactions in quantitative and qualitative traits. Heredity 97:4–18
Jackson CH, Best NG, Richardson S (2009) Bayesian graphical models for regression on multiple data sets with different variables. Biostatistics 10:335–351
Jeffreys H (1961) Theory of probability. Clarendon Press, Oxford
Kabán A (2007) On Bayesian classification with Laplace priors. Patt Rec Lett 28:1271–1282
Kao C-H, Zeng Z-B, Teasdale RD (1999) Multiple interval mapping for quantitative trait loci. Genetics 152:1203–1216
Kass RE, Raftery AE (1995) Bayes factors. J Am Stat Assoc 90:773–795
Knürr T, Läärä E, Sillanpää MJ (2011) Genetic analysis of complex traits via Bayesian variable selection: the utility of a mixture of uniform priors. Genet Res 93:303–318
Kärkkäinen HP, Sillanpää MJ (2012) Back to basics for Bayesian model building in genomic selection. Genetics 191:969–987
Lande R, Thompson R (1990) Efficiency of marker assisted selection in the improvement of quantitative traits. Genetics 124:743–756
Li Y, Campbell C, Tipping ME (2002) Bayesian automatic relevance determination algorithms for classifying gene expression data. Bioinformatics 18:1332–1339
Li Z, Sillanpää MJ (2012a) Estimation of quantitative trait locus effects with epistasis by variational Bayes algorithms. Genetics 190:231–249
Li Z, Sillanpää MJ (2012b) Overview of LASSO-related penalized regression methods for quantitative trait mapping and genomic selection. Theor Appl Genet 125:419–435
Lowd D, Shamaei A (2011) Mean field inference in dependency networks: an empirical study. In: Proceedings of the 25th conference on artificial intelligence (AAAI-11), San Francisco, CA
Lunn D, Best N, Spiegelhalter D, Graham G, Neuenschwander B (2009) Combining MCMC with ‘sequential’ PKPD modelling. J Pharmacokinet Pharmacodyn 36:19–38
Makhuvha T, Pegram G, Sparks R, Zucchini W (1997) Patching rainfall data using regression methods. 1. Best subset selection, EM and pseudo-EM methods: theory. J Hydrol 198:289–307
Malo N, Libiger O, Schork NJ (2008) Accommodating linkage disequilibrium in genetic-association analyses via ridge regression. Am J Hum Genet 82:375–385
McLachlan GJ, Krishnan T (1997) The EM algorithm and extensions. Wiley, New York
Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Mutshinda CM, O’Hara RB, Woiwod IP (2011) A multispecies perspective on ecological impacts of climatic forcing. J Anim Ecol 80:101–107
Mutshinda CM, Sillanpää MJ (2011) Bayesian shrinkage analysis of QTLs under shape-adaptive shrinkage priors, and accurate re-estimation of genetic effects. Heredity 107:405–412
Mutshinda CM, Sillanpää MJ (2010) Extended Bayesian LASSO for multiple quantitative trait loci mapping and unobserved phenotype prediction. Genetics 186:1067–1075
Mutshinda CM, O’Hara RB, Woiwod IP (2009) What drives community dynamics? Proc R Soc B 276:2923–2929
Miller A (2002) Subset selection in regression. Chapman and Hall, London
Myers RL (1992) Classical and modern regression analysis, 2nd edn. Wiley, New-York
O’Hara RB, Sillanpää MJ (2009) A review of Bayesian variable selection methods: what, how and which. Bayesian Anal 4:85–118
R Development Core Team (2011) R: A language and environment for statistical computing, reference index version 2.13.2. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0. http://www.R-project.org
Sen S, Churchill GA (2001) A statistical framework for quantitative trait mapping. Genetics 159:371–387
Shepherd R, Meuwissen THE, Woolliams JA (2010) Genomic selection and complex trait prediction using a fast EM algorithm applied to genome-wide markers. BMC Bioinform 11:529
Sillanpää MJ, Hoti F (2007) Mapping quantitative trait loci from a single tail sample of the phenotype distribution including survival data. Genetics 177:2361–2377
Sillanpää MJ, Bhattacharjee M (2006) Association mapping of complex trait loci with context-dependent effects and unknown context-variable. Genetics 174:1597–1611
Sillanpää MJ, Bhattacharjee M (2005) Bayesian association-based fine mapping in small chromosomal segments. Genetics 169:427–439
Sillanpää MJ, Corander J (2002) Model choice in gene mapping: what and why. Trends Genet 18:301–307
Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989
Sun W, Ibrahim JG, Zou F (2010) Genome-wide multiple loci mapping in experimental crosses by the iterative penalized regression. Genetics 185:349–359
ter Braak CJF, Boer MP, Bink MCAM (2005) Extending Xu’s Bayesian model for estimating polygenic effects using markers of the entire genome. Genetics 170:1435–1438
Tibshirani R (1996) Regression shrinkage and selection via LASSO. J Roy Stat Soc B 58:267–288
Tinker NA, Mather DE, Rosnagel BG, Kasha KJ, Kleinhofs A (1996) Regions of the genome that affect agronomic performance in two-row barley. Crop Sci 36:1053–1062
Tipping ME, Lawrence ND (2005) Variational inference for Student-t models: robust Bayesian interpolation and generalized component analysis. NeuroComputing 69:123–141
Tipping ME (2001) Sparse Bayesian learning and the relevance vector machine. J Mach Learn Res 1:211–244
Wang S, Basten CJ, Zeng Z-B (2006) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC
Wang H, Zhang Y-M, Li X, Masinde GL, Mohan S, Baylink DJ, Xu S (2005) Bayesian shrinkage estimation of quantitative trait loci parameters. Genetics 170:465–480
Whittaker JC, Thompson R, Denham MC (2000) Marker-assisted selection using ridge regression. Genet Res 75:249–252
Xu S (2010) An expectation-maximization algorithm for the Lasso estimation of quantitative trait locus effects. Heredity 105:483–494
Xu S (2007) An empirical Bayes method for estimating epistatic effects of quantitative trait loci. Biometrics 63:513–521
Xu S (2003) Estimating polygenic effects using markers of the entire genome. Genetics 163:789–801
Xu S, Jia Z (2007) Genomewide analysis of epistatic effects for quantitative traits in barley. Genetics 175:1955–1963
Yi N, Banerjee S (2009) Hierarchical generalized linear models for multiple quantitative trait locus mapping. Genetics 181:1101–1113
Yi N, Xu S (2008) Bayesian LASSO for quantitative trait loci mapping. Genetics 179:1045–1055
Yi N, George V, Allison DB (2003) Stochastic search variable selection for identifying multiple quantitative trait loci. Genetics 164:1129–1138
Yi N, Shriner D, Banerjee S, Mehta T, Pomp D, Yandell BS (2007) An efficient Bayes model selection approach for interacting quantitative trait loci models with many effects. Genetics 176:1865–1877
Zielke G (1968) Inversion of modified symmetric matrices. J Assoc Comput Mach 15:402–408
Zou H, Hastie T (2005) Regularization and variable selection via the elastic net. J Roy Stat Soc B 67:301–320
Acknowledgments
The authors wish to thank Hanni Kärkkäinen, Zitong Li and two anonymous referees for their pertinent comments and suggestions. This work was supported by a research grants from the Academy of Finland and University of Helsinki’s research funds.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by F. van Eeuwijk.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mutshinda, C.M., Sillanpää, M.J. Swift block-updating EM and pseudo-EM procedures for Bayesian shrinkage analysis of quantitative trait loci. Theor Appl Genet 125, 1575–1587 (2012). https://doi.org/10.1007/s00122-012-1936-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-012-1936-1