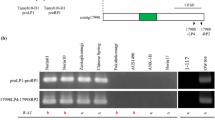
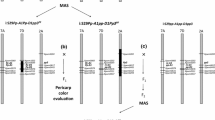
Abstract
The grain color of wheat affects not only the brightness of flour, but also tolerance to preharvest sprouting. Grain color is controlled by dominant R-1 genes located on the long arm of hexaploid wheat chromosomes 3A, 3B, and 3D (R-A1, R-B1, and R-D1, respectively). The red pigment of the grain coat is composed of catechin and proanthocyanidin (PA), which are synthesized via the flavonoid biosynthetic pathway. We isolated the Tamyb10-A1, Tamyb10-B1, and Tamyb10-D1 genes, located on chromosomes 3A, 3B, and 3D, respectively. These genes encode R2R3-type MYB domain proteins, similar to TT2 of Arabidopsis, which controls PA synthesis in testa. In recessive R-A1 lines, two types of Tamyb10-A1 genes: (1) deletion of the first half of the R2-repeat of the MYB region and (2) insertion of a 2.2-kb transposon belonging to the hAT family. The Tamyb10-B1 genes of recessive R-B1 lines had 19-bp deletion, which caused a frame shift in the middle part of the open reading frame. With a transient assay using wheat coleoptiles, we revealed that the Tamyb10 gene in the dominant R-1 allele activated the flavonoid biosynthetic genes. We developed PCR-based markers to detect the dominant/recessive alleles of R-A1, R-B1, and R-D1. These markers proved to be correlated to known R-1 genotypes of 33 varieties except for a mutant with a single nucleotide substitution. Furthermore, double-haploid (DH) lines derived from the cross between red- and white-grained lines were found to necessarily carry functional Tamyb10 gene(s). Thus, PCR-based markers for Tamyb10 genes are very useful to detect R-1 alleles.








Similar content being viewed by others
References
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15(1):63–78
Ahmed N, Maekawa M, Utsugi S, Himi E, Ablet H, Rikiishi K, Noda K (2003) Transient expression of anthocyanin in developing wheat coleoptile by maize C1 and B-peru regulatory genes for anthocyanin synthesis. Breed Sci 53(1):29–34
Ahmed N, Maekawa M, Utsugi S, Rikiishi K, Ahmad A, Noda K (2006) The wheat Rc gene for red coleoptile colour codes for a transcriptional activator of late anthocyanin biosynthesis genes. J Cereal Sci 44:54–58
Akagi T, Ikegami A, Tsujimoto T, Kobayashi S, Sato A, Kono A, Yonemori K (2009) DkMyb4 is a myb transcription factor involved in proanthocyanidin biosynthesis in persimmon fruit. Plant Physiol 151(4):2028–2045
Akagi T, Ikegami A, Yonemori K (2010) DkMyb2 wound-induced transcription factor of persimmon (Diospyros kaki Thunb.), contributes to proanthocyanidin regulation. Planta 232(5):1045–1059
Akiyama Y, Onizuka K, Nogushi T, Ando M (1998) Parallel protein information analysis (papia) system running on a 64-node pc cluster. In: Proc the 9th genome informatics workshop (GIW’98), 1998. Universal Academy Press, pp 131–140
Allan RE, Vogel OA (1965) Monosomic analysis of red seed color in wheat. Crop Sci 5:474–475
Bender J, Fink GR (1998) A Myb homologue, ATR1, activates tryptophan gene expression in Arabidopsis. Proc Natl Acad Sci USA 95(10):5655–5660
Biffen RH (1905) Mendel’s laws of inheritance and wheat breeding. J Agric Sci 1:4–48
Bogs J, Jaffe FW, Takos AM, Walker AR, Robinson SP (2007) The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol 143(3):1347–1361
Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12(12):2383–2394
Buck MJ, Atchley WR (2003) Phylogenetic analysis of plant basic helix–loop–helix proteins. J Mol Evol 56(6):742–750
Choulet F, Wicker T, Rustenholz C, Paux E, Salse J, Leroy P, Schlub S, Le Paslier MC, Magdelenat G, Gonthier C, Couloux A, Budak H, Breen J, Pumphrey M, Liu S, Kong X, Jia J, Gut M, Brunel D, Anderson JA, Gill BS, Appels R, Keller B, Feuillet C (2010) Megabase level sequencing reveals contrasted organization and evolution patterns of the wheat gene and transposable element spaces. Plant Cell 22(6):1686–1701
Debeaujon I, Leon-Kloosterziel KM, Koornneef M (2000) Influence of the testa on seed dormancy, germination, and longevity in arabidopsis. Plant Physiol 122(2):403–414
Flintham JE (2000) Different genetic components control coat-imposed and embryo-imposed dormancy in wheat. Seed Sci Res 10:43–50
Grotewold E, Drummond BJ, Bowen B, Peterson T (1994) The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76(3):543–553
Himi E, Noda K (2004) Isolation and location of three homoeologous dihydroflavonol-4-reductase (DFR) genes of wheat and their tissue-dependent expression. J Exp Bot 55(396):365–375
Himi E, Noda K (2005) Red grain colour gene (R) of wheat is a myb-type transcription factor. Euphytica 143:239–242
Himi E, Mares DJ, Yanagisawa A, Noda K (2002) Effect of grain colour gene (R) on grain dormancy and sensitivity of the embryo to abscisic acid (ABA) in wheat. J Exp Bot 53(374):1569–1574
Himi E, Nisar A, Noda K (2005) Colour genes (R and Rc) for grain and coleoptile upregulate flavonoid biosynthesis genes in wheat. Genome 48(4):747–754
Huang J, Zhang K, Shen Y, Huang Z, Li M, Tang D, Gu M, Cheng Z (2009) Identification of a high frequency transposon induced by tissue culture, nDaiZ, a member of the hAT family in rice. Genomics 93(3):274–281
Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19(22):6150–6161
Kaneko S, Komae K, Nagamine T, Yamada T (1994) Development of an extraction procedure for wheat red coat pigments and determination of varietal differences for this trait. Breed Sci 44:263–266
Kempken F, Windhofer F (2001) The hAT family: a versatile transposon group common to plants, fungi, animals, and man. Chromosoma 110(1):1–9
Khlestkina EK, Roder MS, Salina EA (2008) Relationship between homoeologous regulatory and structural genes in allopolyploid genome—a case study in bread wheat. BMC Plant Biol 8:88
Koval SF (1997) The catalog of near-isogenic lines of novosibirskaya-67 common wheat and principles of their use in experiments. Russ J Genetics 33(8):995–1000
Kristiansen KN, Rohde W (1991) Structure of the Hordeum vulgare gene encoding dihydroflavonol-4-reductase and molecular analysis of ant18 mutants blocked in flavonoid synthesis. Mol Gen Genet 230(1–2):49–59
Lee MM, Schiefelbein J (1999) WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99(5):473–483
Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57:405–430
Mares D, Mrva K, Cheong J, Williams K, Watson B, Storlie E, Sutherland M, Zou Y (2005) A QTL located on chromosome 4A associated with dormancy in white- and red-grained wheats of diverse origin. Theor Appl Genet 111(7):1357–1364
Mares D, Rathjen J, Mrva K, Cheong J (2009) Genetic and environmental control of dormancy in white-grained wheat (Triticum aestivum L.). Euphytica 168:311–318
Matus-Cadiz MA, Daskalchuk TE, Verma B, Puttick D, Chibbar RN, Gray GR, Perron CE, Tyler RT, Hucl P (2008) Phenolic compounds contribute to dark bran pigmentation in hard white wheat. J Agric Food Chem 56(5):1644–1653
McCallum JA, Walker JRL (1990) Proanthocyanidins in wheat bran. Cereal Chem 67(3):282–285
McCarty DR, Hattori T, Carson CB, Vasil V, Lazar M, Vasil IK (1991) The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66:895–905
McIntosh RA, Hart GE, Devos KM, Gale MD, Rogers WJ (1998) Catalogue of gene symbols for wheat. In: Proceedings of the ninth international wheat genetics symposium, Canada, 1998. University of Saskatchewan Extension Press
Mellway RD, Tran LT, Prouse MB, Campbell MM, Constabel CP (2009) The wound-, pathogen-, and ultraviolet B-responsive MYB134 gene encodes an R2R3 MYB transcription factor that regulates proanthocyanidin synthesis in poplar. Plant Physiol 150(2):924–941
Metzger RJ, Silbaugh BA (1970) Location of genes for seed coat color in hexaploid wheat, Triticum aestivum L. Crop Sci 10:495–496
Miura H, Sato N, Kato K, Amano Y (2002) Detection of chromosomes carrying genes for seed dormancy of wheat using the backcross reciprocal monosomic method. Plant Breed 121:394–399
Miyamoto T, Everson EH (1958) Biochemical and physiological studies of wheat seed pigmentation. Agron J 50:733–734
Mol J, Grotewold E, Koes R (1998) How genes paint flowers and seeds. Trends Plant Sci 3(6):212–217
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8(19):4321–4325
Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13(9):2099–2114
Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD (1991) A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67(3):483–493
Quinde-Axtell Z, Baik BK (2006) Phenolic compounds of barley grain and their implication in food product discoloration. J Agric Food Chem 54(26):9978–9984
Ramsay NA, Glover BJ (2005) MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10(2):63–70
Routaboul JM, Kerhoas L, Debeaujon I, Pourcel L, Caboche M, Einhorn J, Lepiniec L (2006) Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 224(1):96–107
Sears ER (1944) Cytogenetic studies with polyploid species of wheat. II. Additional chromosomal aberrations in Triticum vulgare. Genetics 29:232–246
Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4(5):447–456
Styles ED, Ceska O (1975) Genetic control of 3-hydroxy- and 3-deoxy-flavonoid in Zea mays. Phytochemistry 14:413–415
Suzuki A, Suzuki T, Tanabe F, Toki S, Washida H, Wu CY, Takaiwa F (1997) Cloning and expression of five myb-related genes from rice seed. Gene 198(1–2):393–398
Terrier N, Torregrosa L, Ageorges A, Vialet S, Verries C, Cheynier V, Romieu C (2009) Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol 149(2):1028–1041
Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K (1993) An Arabidopsis myb homolog is induced by dehydration stress, and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5(11):1529–1539
Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11(7):1337–1350
Warner RL, Kudrna DA, Spaeth SC, Jones SS (2000) Dormancy in white-grain mutants of Chinese Spring wheat (Triticum aestivum L.). Seed Sci Res 10:51–60
Winkel-Shirley B (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126(2):485–493
Xie DY, Jackson LA, Cooper JD, Ferreira D, Paiva NL (2004) Molecular and biochemical analysis of two cDNA clones encoding dihydroflavonol-4-reductase from Medicago truncatula. Plant Physiol 134(3):979–994
Yamada T (1990) Classification of GA response, Rht genes and culm length in Japanese varieties and landraces of wheat. Euphytica 50:221–239
Yoshida K, Iwasaka R, Kaneko T, Sato S, Tabata S, Sakuta M (2008) Functional differentiation of Lotus japonicus TT2s, R2R3-MYB transcription factors comprising a multigene family. Plant Cell Physiol 49(2):157–169
Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like bHLH proteins. Plant J 40(1):22–34
Acknowledgments
We thank Dr. Flintham (John Innes Centre) for providing Novosibirskaya 67 and its NIL; Dr. Mares (The University of Adelaide) for the gift of AUS1490 and EMS-AUS; Dr. Sorrells (Cornell University) for giving Cornell 595; and Dr. Tanio (National Agricultural Research Center, Japan) for providing SZF. This work was supported by grants from the Iijima Memorial Foundation for the Promotion of Food Science and Technology and the Ministry of Agriculture, Forestry, and Fisheries of Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Sorrells.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2011_1555_MOESM1_ESM.ppt
Fig. S1 Positions of primers in this study. Three exons of Tamyb10 gene are described with rectangles. Black-painted regions represent the MYB consensus region. Untranslated regions and introns are described with horizontal lines. Primers are shown with triangles. Right-directed triangles indicate sense primers, and left-directed triangles indicate antisense primers. Primers shown with white triangles represent the primers designed on the basis of the maize P gene. Primers shown with black triangles are designed on the basis of Tamyb10 genes. (PPT 112 kb)
122_2011_1555_MOESM2_ESM.ppt
Fig. S2 Genome-determined primers for Tamyb10. a Fragments of the Tamyb10-A1 gene were amplified with Tamyb10-LP1 and Tamyb10-RP1. b Fragments of the Tamyb10-B1 gene were amplified with Tamyb10-LP2 and Tamyb10-RP1. c Fragments of the Tamyb10-D1 gene were amplified with Tamyb10-LP3 and Tamyb10-RP3. (PPT 126 kb)
122_2011_1555_MOESM3_ESM.ppt
Fig. S3 Primers and PCR conditions for Tamyb10 amplification. PCR conditions are described in boxes below the structures of each Tamyb10 gene. The expected sizes of amplification fragments using each set of primers are listed in the tables. a-c Tamyb10-A1 genes of Norin 61 (R-A1b), CS (R-A1a), and Norin 17 (R-A1a). The red and black boxes indicate exons. The arrows indicate primer positions. A grey region with a black box in Tamyb10-A1 of CS represents an unknown sequence. 2.2 kb of GeST was inserted into second intron of Tamyb10-A1 of Norin 17. b Tamyb10-B1 gene of CS (R-B1a). The green and black boxes indicate exons. A white triangle represents the site of a 19-bp deletion. Amino acid sequences are altered by a frame shift. c Tamyb10-D1 gene of CS (R-D1b). The blue boxes indicate exons.(PPT 462 kb)
122_2011_1555_MOESM4_ESM.ppt
Fig. S4 Deduced amino acid sequences encoded by Tamyb10-A1 of AUS1490 (R-A1b), Tamyb10-B1 of Norin 61 (R-B1b), and Tamyb10-D1 of CS (R-D1b). The R2 and R3 repeats of the MYB consensus region are shaded in black and gray, respectively. Tamyb10-A1 of CS (R-A1a) lacked the N-terminal sequences shown in blue letters of Tamyb10-A1 of AUS1490 (R-A1b). A white rectangle indicates the substituted residue from glycine (G) to glutamic acid (E) in EMS-AUS. A white triangle indicates the 19-bp deletion site of Tamyb10-B1 in CS. Essential residues of the conserved amino acid signature (D/ELx2R/Kx3Lx6Lx3R) as the structural basis for interaction between MYB and bHLH proteins are shown with a dotted black circle. The conserved sequences (IRTKAL/IRC) among Tamyb10, Arabidopsis TT2, and rice OsMYB3 are shown in bold letters, and the KAxRC sequences in the motif also seen in C1 protein are shown in red letters. (PPT 124 kb)
Rights and permissions
About this article
Cite this article
Himi, E., Maekawa, M., Miura, H. et al. Development of PCR markers for Tamyb10 related to R-1, red grain color gene in wheat. Theor Appl Genet 122, 1561–1576 (2011). https://doi.org/10.1007/s00122-011-1555-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-011-1555-2