Abstract
Asian soybean rust (ASR), caused by Phakopsora pachyrhizi Syd., has the potential to become a serious threat to soybean, Glycine max L. Merr., production in the USA. A novel rust resistance gene, Rpp?(Hyuuga), from the Japanese soybean cultivar Hyuuga has been identified and mapped to soybean chromosome 6 (Gm06). Our objectives were to fine-map the Rpp?(Hyuuga) gene and develop a high-throughput single nucleotide polymorphism (SNP) assay to detect this ASR resistance gene. The integration of recombination events from two different soybean populations and the ASR reaction data indicates that the Rpp?(Hyuuga) locus is located in a region of approximately 371 kb between STS70887 and STS70923 on chromosome Gm06. A set of 32 ancestral genotypes which is predicted to contain 95% of the alleles present in current elite North American breeding populations and the sources of the previously reported ASR resistance genes (Rpp1, Rpp2, Rpp3, Rpp4, Rpp5, and rpp5) were genotyped with five SNP markers. We developed a SimpleProbe assay based on melting curve analysis for SNP06-44058 which is tighly linked to the Rpp?(Hyuuga) gene. This SNP assay can differentiate plants/lines that are homozygous/homogeneous or heterozygous/heterogeneous for the resistant and susceptible alleles at the Rpp?(Hyuuga) locus.
Similar content being viewed by others
Introduction
Asian soybean rust (ASR) caused by Phakopsora pachyrhizi Syd. is a widespread disease of soybean [Glycine max (L.) Merr.] and has the potential to cause serious economic losses in soybean production (Sinclair and Hartman 1996; Grau et al. 2004). The pathogen was recently reported for the first time in North America (Schneider et al. 2005). Researchers during the 1970s and 1980s identified four unique dominant ASR resistance genes. PI 200492 (‘Komata’) has a single dominant gene (Rpp1) that confers resistance to an Australian Asian rust isolate (McLean and Byth 1980; Bromfield and Hartwig 1980). PI 230970 (‘Ankur’) has Rpp2 (Singh and Thapliyal 1977), and PI 462312 has the ASR resistance gene Rpp3 (Hartwig and Bromfield 1983). The cultivar Bing Nan (PI 459025) from China possesses a dominant resistance allele (Rpp4) at a locus different from the other three resistance alleles (Hartwig 1986). Recently, a new dominant resistance allele (Rpp5) and a recessive resistance allele (rpp5) were reported by Garcia et al. (2008). A novel rust resistance gene, Rpp?(Hyuuga) from the Japanese cultivar Hyuuga, produces a red-brown (RB) or resistant lesion type (LT) with current rust isolates prevalent in the USA. The resistance gene was identified and mapped to a 3.5 cM region on soybean chromosome Gm06 [linkage group (LG) C2] (Monteros et al. 2007). The use of native Asian soybean rust resistance genes should be an important management component for the disease, but to date no commercial cultivars possessing any ASR resistance genes are currently available in the USA.
Strategically, DNA markers are used to identify genetic factors for traits of interest and to introgress them more efficiently into elite cultivars than can be accomplished using traditional breeding approaches. Single base changes in sequence, called single nucleotide polymorphisms (SNPs), are the most abundant source of variation in plant genomes and include small insertions and deletions (indels) (Zhu et al. 2003). Compared to simple sequence repeats (SSRs), SNPs are more amenable to high-throughput automated genotyping assays that allow samples to be genotyped faster and more economically than with SSRs (Hurley et al. 2004; Ha and Boerma 2008). SNP detection technologies have evolved from expensive, time consuming, and labor intensive processes to some of the most highly automated, efficient, and relatively inexpensive methods of DNA marker detection (Kwok and Chen 2003).
Soybean has a relatively low sequence variation due to historical genetic bottlenecks and narrow genetic base (Hyten et al. 2006; Gizlice et al. 1993; Carter et al. 2004). A combination of soybean ancestors and first progeny that contribute at least 95% of the alleles found in public cultivars in North America from 1947 to 1988 has been identified (Gizlice et al. 1994). Fourteen genotypes estimated to have contributed 80.5% of the allelic diversity present in North American soybean cultivars were used to determine the SNP frequency in coding and non-coding soybean DNA sequence (Gizlice et al. 1994; Zhu et al. 2003). Soybean has, on average, about 1.95 SNPs per 534 bp and over 1,100 single nucleotide polymorphism (SNP) markers across 20 soybean chromosomes have been mapped (Choi et al. 2007). A SNP haplotype refers to a distinct combination of SNPs that are tightly linked in a region of a chromosome or blocks of DNA which tend to be inherited as entire units from a parent to its progeny (Shastry 2004). SNPs that differentiate one haplotype from another are potentially useful as markers linked to QTL or genes and can be used for marker-assisted selection (MAS). Individuals susceptible to a disease and having a shared haplotype can be grouped together, and the haplotype can be used to describe other individuals that will likely be susceptible to a disease. The use of MAS allows selection to be performed in early generations at early stages of plant development and in the absence of the pest or pathogen (Mohan et al. 1997; Ha et al. 2007).
There are many techniques available to genotype SNPs (Kwok and Chen 2003). Homogenous, closed-tube methods for SNP genotyping that do not require a separation step are attractive for their simplicity and containment of amplified products. Most of these methods are based on PCR and use oligonucleotide probes (Liew et al. 2004). High-resolution melting curve analysis without oligonucleotide probes has proven to be a highly sensitive method for mutation discovery and SNP genotyping (Cho et al. 2008). Nucleic acid melting is tracked by monitoring the fluorescence of the samples across a defined temperature range, generating high-resolution melting profiles that can be used to identify the presence of sequence variation within the amplicon (deSilva and Blackett 2007). Wittwer et al. (2003) used high-resolution melting curve analysis of a 110 bp PCR amplicon to genotype the single nucleotide polymorphisms hemoglobin (Hb)C(G16A) and HbS (A17T) near the middle of the amplicon. Another closed-tube method for SNP genotyping is high-resolution melting analysis using an unlabeled probe. In this case, DNA amplified in the presence of the DNA binding saturation dye emits a high level of fluorescence when double stranded but loses fluorescence as the DNA duplex melts apart enabling the detection of sequence variations in the unlabeled probe (Margraf et al. 2008; Zhou et al. 2004).
Unlike high-resolution melting curve analysis, the SimpleProbe system utilizes a single-labeled genotyping probe designed to increase in fluorescence upon target hybridization. SimpleProbes show a decrease in fluorescence as the genotyping probe is melted from its target strand. The genotypes are determined by evaluating the probe-target duplex melting temperature (T m) of the genotyping probe as it melts from a site of variance producing a characteristic DNA duplex melting profile (Bennett et al. 2003). The T m difference between SNP alleles is determined by the probe length and the base mismatch (Liew et al. 2007). Base-pair mismatches shift the stability of a duplex by varying amounts depending on the particular mismatch, the mismatch position, and neighboring base pairs (SantaLucia et al. 1996). The SimpleProbe assay was successfully applied in a soybean marker-assisted breeding program for selecting southern root-knot nematode resistance (Ha and Boerma 2008).
The objectives of this research were to fine map the genomic location of the Rpp?(Hyuuga) locus and to develop a SNP assay using the SimpleProbe system to detect the Rpp?(Hyuuga) ASR resistance gene for use in MAS.
Materials and methods
Plant materials
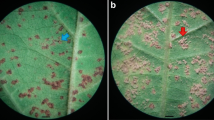
Two populations were used for fine-mapping the Rpp?(Hyuuga) gene in this study. The first population consists of 100 F6-derived recombinant inbred lines (RILs) from the cross of ‘Dillon’ (tan or susceptible lesion type) × Hyuuga (RB or resistant lesion type) (Shipe et al. 1997; GRIN 2009). The second population consists of 92 F6-derived RILs from the cross of ‘Benning’ (tan or susceptible lesion type) × Hyuuga (Boerma et al. 1997). DNA was extracted from the RILs in both populations and they were genotyped with SSR markers (Satt460, Satt079, Staga001, and Satt307) on chromosome Gm06 as described by Monteros et al. (2007). Based on the genotype of four SSR markers and five newly developed SNP markers in this study, six recombinant substitution lines (RSLs) were identified from the Dillon × Hyuuga population (Fig. 1). Three replications of each RIL and the parents, with four plants per replication, were planted in a randomized complete block design in the greenhouse at the University of Georgia’s Griffin campus on 6, October, 2006, to evaluate their reaction to ASR. Plants were inoculated with the ASR spore suspension on 23, October, 2006, according to the procedure described in Monteros et al. (2007). The P. pachyrhizi spores were obtained from ASR-infected soybean plants collected at the University of Georgia’s Attapulgus Research and Education Center, located in the southwest corner of Georgia, near the Georgia–Florida border. Data on lesion type, lesion number, and visible sporulation was obtained on 9, November, 2006.
In the Benning × Hyuuga population, one plant that was heterozygous for the genomic region between markers Satt460 and Satt307 was initially identified from among the F7 plants within a single RIL. After self-pollination of this plant, we obtained 52 F8 seeds and these seeds were planted in the greenhouse. DNA was extracted from individual F8 plants using a modified CTAB procedure described by Keim et al. (1988) and both SSR and SNP markers located in the Satt460 to Satt307 genomic region were used to find recombinant events. Based on their marker genotypes, four plants out of 52 F8 plants were placed into four recombinant classes, Class I–IV (Fig. 2), but they were heterozygous for the genomic region. After self-pollination of the four heterozygous plants we obtained a total of 376 F9 seeds. DNA was extracted from individual seed chips using the procedure described by Edwards et al. (1991). After genotyping with SNP and SSR markers from F9 seeds, we identified homozygous seeds: 12 seeds in Class I, 20 in Class II, 20 in Class III, and 2 in Class IV. To evaluate their reaction to ASR, six seeds each from Class I, II, and III, the two seeds from Class IV and the parents, Hyuuga and Benning, were planted in the greenhouse at the University of Georgia’s Griffin campus on 10, July, 2009. Plants were inoculated twice with the ASR spore suspension on 6, August, 2009 and 10, August, 2009. Data on lesion type, lesion number, and visible sporulation were obtained on 24, August, 2009.
Graphical genotypes of representative RSLs from Benning × Hyuuga in the Rpp?(Hyuuga) region on chromosome Gm06. LT indicates the type of lesion, Tan is the susceptible reaction and RB is the resistant reaction. STS70887, STS70891, STS70919, and STS70923 previously identified by Hyten et al. (2009) were located at nucleotide 43746435, 43786392, 44086038, and 44118117 on Gm06, respectively
Twenty-five seeds of the 32 soybean ancestors and first progeny that contribute at least 95% of the alleles found in public cultivars in North America from 1947 to 1988 (Gizlice et al. 1994) and previously reported sources of ASR resistance genes, PI 200492 (Rpp1); PI 230970 (Rpp2); PI 462312 (Rpp3); PI 459025B (Rpp4); PI 200526 (Rpp5); PI 200487 (Rpp5); PI 471904 (Rpp5); PI 200456 (rpp5); and PI 506764 (Rpp?(Hyuuga)), obtained from the USDA Soybean Germplasm Collection maintained at the University of Illinois (Urbana, IL, USA) were planted in the greenhouse. Trifoliolate leaves from all plants were harvested and bulked, and processed for DNA extraction as previously described by Monteros et al. (2007).
SNP identification
An amplicon resequencing approach was used to identify SNPs associated with Rpp?(Hyuuga). Chromosome Gm06 from 43 to 45 Mbp templates were obtained from the soybean genome sequence database (http://www.phytozome.net/soybean), and a total of 50 primers were designed from this genomic region with the LightCycler Probe Design software (Roche Applied Science, Indianapolis, IN, USA).
All PCR primers were used to amplify genomic DNA of Dillon and Hyuuga. The PCR products were analyzed on a 1.5% agarose gel stained with ethidium bromide. A total of 29 out of 50 primer pairs produced a single amplicon from genomic DNA. These PCR products were directly sequenced using one of the PCR primers with a BigDye Terminator v3.1 Cycle Sequencing Kit (PE-ABI, Foster City, CA, USA). These amplicons were analyzed on an ABI 3730 automated sequencer (PE-ABI, Foster City, CA, USA). The sequence data were analyzed with Sequencher™ 4.2 (GeneCodes Corp., Ann Arbor, MI, USA). Only 12 out of 29 amplicons (41%) contained SNPs or InDels between Dillon and Hyuuga (Table 1). An additional 23 STS markers located between Satt460 and Sat_263 from the study of Hyten et al. (2009) were also evaluated by sequencing as previously described.
SNP genotyping using high-resolution melting analysis with unlabeled probe
The SNP markers identified above were used to genotype the Dillon × Hyuuga RILs, the 32 ancestral genotypes, Dillon, and the nine plant introductions with previously reported ASR resistance genes (Table 4). High-resolution melting analysis with unlabeled oligonucleotide probes targeting SNP sites and a saturation DNA dye was applied in this study. Five SNPs were successfully genotyped using unlabeled probes (Table 2).
All PCR reactions were performed in 384-well plates with a total volume of 3 μL per well and conducted as asymmetry. The PCR reaction mixture consisted of 20–30 ng of genomic DNA, 3 mM MgCl2, 1 μM of excess primer, 0.2 μM of limiting primer, 0.5× of LightCycler 480 Genotyping Master mix (Roche Diagnostics, Indianapolis, IN, USA), and 0.6× LCGreen Plus (Idaho Technology, Salt Lake City, UT, USA). PCR was performed in the Light Cycler® 480 (Roche Applied Science, Indianapolis, IN, USA) for 50 cycles with 10 s denaturation at 95°C, 15 s of annealing at 58°C, and 20 s extension at 72°C. After amplification, 1.0 μM of unlabeled oligonucleotide probe was added and a final melting cycle was performed by raising the temperature to 95°C for 2 min, lowering the temperature to 40°C for 5 min, and increasing the temperature to 90°C with continuous fluorescent acquisition followed by a cool down to 40°C.
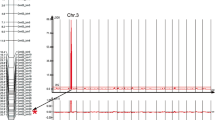
The fluorescence signal (F) was plotted in real time against temperature (T) to produce melting curves for each sample. Melting curves were then converted to negative derivative curves of fluorescence with respect to temperature (−dF/dT) by the LightCycler® Data Analysis software (Roche Diagnostics, Indianapolis, IN, USA). The same software was used to group similar melting curves and automatically call genotypes based on melting standards for known genotypes in the experiment or software-defined melting standards.
Design of a simple probe to detect Rpp?(Hyuuga)
A SNP site from SNP06_44058 was detected at position 44058927 on chromosome Gm06, in which the ASR-susceptible lines had the “G” allele and the ASR-resistant lines had the “T” allele. For melting curve analysis, a 180 bp fragment was amplified using the forward 5′-GGT TTT CAG ACA AGA TCA AAT CG-3′ and reverse primers 5′-CCA TTC ATA GTG AAC TCA AC-3′. The SimpleProbe 5′-CTG AAG GGT XI CAA AGT GCC AGA TC-3′ was internally labeled with fluorescein and blocked with phosphate at the 3′ end (TIB MOLBIOL, Adelphia, NJ, USA). The probe was designed to match the resistant allele.
PCR reactions were performed in 384-well plates with a total volume of 3 μL per well in a LightCycler 480 (Roche Applied Science, Indianapolis, IN, USA), following the standard protocol for asymmetrical PCR. The PCR reaction mixture consisted of 20–30 ng of genomic DNA, 0.5 μM of forward primer, 1.0 μM of reverse primer, 0.2 μM of SimpleProbe, 3.0 mM MgCl2, and 0.5× of LightCycler 480 Genotyping Master mix.
After an initial denaturation of 5 min at 95°C, 50 PCR cycles were performed with 10 s of denaturation at 95°C, 15 s of annealing at 55°C, and 20 s extension at 72°C. A final melting cycle was performed by raising the temperature to 95°C for 2 min, lowering the temperature to 40°C for 2 min, and increasing the temperature to 90°C, at this point using continuous fluorescent acquisition, and followed by a final cool down to 40°C. The data from the Light Cycler® 480 was analyzed as described earlier.
Results
The Dillon × Hyuuga RILs and the RSLs developed from Benning × Hyuuga were genotyped with SSR and SNP markers to fine-map the Rpp?(Hyuuga) gene. The Rpp?(Hyuuga) locus was previously mapped on chromosome Gm06 (LG-C2) in the 3.5 cM interval between the SSR markers Satt460 and Satt307 (Monteros et al. 2007). Two additional SSR markers, Satt079 and Staga001, were mapped between Satt460 and Satt307. Resequencing of the amplicons containing the SNP site was used based on the availability of the soybean whole genome shotgun assembly (http://www.phytozome.net/soybean). Sequence information from the four SSR markers was used in BLAST searches against the soybean genomic sequence database. The SSR markers Satt460, Satt079, Staga001, and Satt307 were physically placed at approximately 43291288, 43950925, 45427178, and 46286824 on chromosome Gm06, respectively (Table 1). Of the 50 PCR primers designed to amplify 2,135 bp of sequence located in the Satt460-Staga001 interval from Dillon and Hyuuga, only 29 of those primers produced a single band on agarose gels which was then used for sequencing. Twelve out of 29 PCR fragments contained sequence variation between Dillon and Hyuuga and produced a sequence tagged site (STS) for fine-mapping of Rpp?(Hyuuga). A total of 77 SNPs and 8 InDels were identified in these 12 STS. We used high-resolution melting analysis with unlabeled probe to genotype the STS. One SNP for each of the 12 STS was selected and used to design single oligonucleotide probes to anneal with the DNA from either Dillon or Hyuuga. Five probes showed clearly different melting curves between Dillon and Hyuuga (Table 2).
These five SNP markers and four SSR markers covering 3 Mbp on chromosome Gm06 were used to screen RILs from Dillon × Hyuuga. A total of six RILs, four of which contain different recombinant events in the target region, were identified and inoculated with ASR (Fig. 1). The RIL 256, 341, and 248 were susceptible to ASR, while RILs 33, 323, and 12 were resistant to ASR based on lesion type. RIL 248 is homozygous for the Dillon genome in the interval between Satt460 and SNP06-44421 and has a tan or susceptible ARS phenotype. RIL 33 is homozygous for the Dillon allele at SNP06-43621 but homozygous for Hyuuga allele at the SNP06-43731, and has a RB or resistant phenotype. The integration of recombination events and the ASR reaction data indicate that the Rpp?(Hyuuga) locus is located between SNP06-43621 and Staga001 on chromosome Gm06.
Additional recombinant events between SNP06-43621 and Staga001 were identified in the Benning × Hyuuga RSLs. After identification of the four initial recombinant classes, more STS markers identified by Hyten et al. (2009) were used to further refine the genomic location of recombination events. Plants in Class I contain a recombinant event between STS70919 and STS70923 and have a tan or susceptible ARS phenotype, while Class II plants contain a recombinant event between STS70887 and STS70891 and have a RB or resistant phenotype. These data suggested that Rpp?(Hyuuga) is located in the region of approximately 371 kb between STS70887 at nucleotide 43746435 and STS70923 at nucleotide 44118117. Seven genes showing a similarity with NBS-LRR disease resistance proteins were annotated in this 371 kb sequence from the Williams 82 genomic sequence (Soybean Genome Project, DoE Joint Genome Institute) (Table 3).
The SNP genotypes for the 32 ancestral lines, Dillon, Hyuuga, and the other eight previously reported sources of ASR resistance genes at five SNP markers are shown in Table 4. All 32 ancestral soybean accessions and Dillon possess a G allele at nucleotide 44058927 (SNP06-44058) on chromosome Gm06, while Hyuuga contains a T allele. ‘Arksoy’, ‘Ralsoy’, and ‘Mejiro’ showed the same SNP genotypes as Hyuuga at the other four markers, SNP06-43621, SNP06-43731, SNP06-43983, and SNP06-44421. Hyuuga, PI 462312 (Rpp3), PI 200487 (Rpp5), and PI 471904 (Rpp5) showed the same SNP genotypes for all five SNPs. The SNP marker SNP06-44058 can be used to distinguish the Rpp?(Hyuuga) allele from the rpp?(Hyuuga) allele in these 32 ancestral genotypes and the previously reported sources of ASR resistance other than PI 462312, PI 200487, and PI 471904. We developed melting curve analysis with SimpleProbe chemistry for use in MAS for Rpp?(Hyuuga). The plants containing the Hyuuga allele yielded melting peaks at 60°C, and the plants containing the Dillon allele yielded melting peaks at 54°C (data not shown). The probe annealed with the Hyuuga sequence and resulted in a 6°C higher T m of the probe with the target sequence compared with the T m of the probe with the mismatch in the Dillon sequence. The heterozygous samples generated both peaks. This assay was further used to genotype the two parents and 100 RILs from the cross of Dillon × Hyuuga. Linkage analysis from the 100 RILs from the cross of Dillon × Hyuuga mapped the SNP marker to the interval between Satt079 and Staga001 (data not shown). The location of the SNP marker from the genetic mapping analysis is in agreement with its physical location on chromosome Gm06.
Discussion
The susceptibility of currently available elite soybean cultivars to ASR and the presence of ASR in most soybean-growing regions in the world make the development of ASR-resistant soybean cultivars an important breeding objective. The detection of additional SNP and SSR markers in the 3.5 cM interval on chromosome Gm06 where Rpp?(Hyuuga) was previously mapped allowed us to increase the resolution in mapping this resistance gene. A similar approach of combining the genotypes of RSLs with their phenotypic response to defoliating insects was used by Zhu et al. (2006) to fine map a major insect resistance QTL on chromosome Gm07 (LG-M). A higher density of markers in this region of chromosome Gm06 reduces the likelihood of recombination between markers and the target loci during the breeding process and should increase the feasibility of fine mapping and map-based cloning of this ASR resistance gene.
In this study, an increase in the marker density allowed us to identify RSLs in the region of the Rpp?(Hyuuga) locus. The integration of the SNP genotypes from selected RSLs and the ASR reaction data allowed us to narrow down the location of the Rpp?(Hyuuga) locus to a 371 kb segment on chromosome Gm06. Based on the gene annotation in the Soybean Genome Project, seven candidate genes containing TIR-NBS-LRR groups were identified in the sequence between STS70887 and STS70923 on chromosome Gm06 (Table 3). These results should increase the feasibility of cloning the Rpp?(Hyuuga) gene following a similar approach used recently by Meyer et al. (2009) to identify the Rpp4 candidate gene from the resistant genotype PI459025B. They found that this candidate gene is a member of the CC-NBS-LRR family of disease resistance genes. This report suggests that other ASR genes may also belong to the NBS-LRR family of disease resistance genes.
Five SNP markers produce a distinct haplotype which can be used to distinguish Hyuuga from other reported sources of ARS resistance genes including Rpp1, Rpp2, Rpp4, Rpp5 from PI 200526, and rpp5, as well as from the soybean ancestors evaluated which contribute a high percent of the genetic variation to elite soybean breeding populations in the USA (Table 4). Both the Rpp1 and the Rpp4 loci have been mapped to chromosome Gm18 (LG-G), and the Rpp2 locus has been mapped to chromosome Gm16 (LG-J) using SSR markers (Hyten et al. 2007; Silva et al. 2008) indicating that Rpp?(Hyuuga) is different from Rpp1, Rpp2, and Rpp4. In our study the sources of Rpp3 (PI 462312) and Rpp5 (PI 200487 and PI 471904) had the same genotype at the five SNP markers as Hyuuga. Hyten et al. (2009) used a F2:3 population from the cross of Williams 82 × PI 462312 to map the Rpp3 gene to a similar region on chromosome Gm06 as Rpp?(Hyuuga). They also found that Hyuuga (PI 506764) and PI 463312 had the same SNP genotypes at 273 of 275 SNPs in this region. However, when PI 462312 and Hyuuga were simultaneously evaluated using Brazilian isolates of ASR, PI 462312 showed a susceptible tan lesion type while Hyuuga produced the resistant RB lesion type (Garcia et al. 2008). Also, Rpp3 was defeated by the P. pachyrhizi MT isolate just 2 years after ASR was first detected in Brazil (Pierozzi et al. 2008). Current efforts include screening Hyuuga and PI 462312 with multiple ASR isolates in Bio-Safety Level 3 containment facilities at Ft. Detrick to identify an isolate of P. pachyrhizi that differentiates Rpp?(Hyuuga) from Rpp3 (Reid Fredrick, Fort Detrick MD, personal communication).
Although Rpp?(Hyuuga) was identified in a Dillon × Hyuuga population, SSR markers near this ASR resistance loci correctly predicted the ASR-resistant reaction in progeny from a Benning × Hyuuga population (Monteros et al. 2007) suggesting that this resistance gene loci can be effective in different genetic backgrounds. The development of a high-throughput SNP-based marker enables breeders in a wide range of soybean breeding programs to use MAS to select for the Rpp?(Hyuuga) ASR resistance gene. The application of the melting curve assay for high-throughput and rapid SNP genotyping in soybean was described by Ha and Boerma (2008). One limitation of using linked markers in MAS is that often a given marker is not polymorphic across all populations (Xu and Crouch 2008). Having a rapid SNP assay that differentiates the Rpp?(Hyuuga) resistance allele from the rpp?(Hyuuga) allele in most elite US breeding populations greatly enhances its utility and efficiency for MAS.
Our newly developed SimpleProbe assay can detect alternate alleles at the Rpp?(Hyuuga) locus and will allow breeders to effectively select ASR-resistant progeny in early segregating generations and facilitate breeding efforts to pyramid Rpp?(Hyuuga) with other ASR resistance genes. These strategies should accelerate the development of ARS-resistant soybean cultivars adapted to the various production regions of the USA.
References
Bennett CD, Campbell MN, Cook CJ, Eyre DJ, Nay LM, Nielsen DR, Rasmussen RP, Bernard PS (2003) The LightTyper™: high-throughput genotyping using fluorescent melting curve analysis. BioTechniques 34:1288–1295
Boerma HR, Hussey RS, Phillips DV, Wood ED, Rowan GB, Finnerty SL (1997) Registration of ‘Benning’ soybean. Crop Sci 37:1982
Bromfield KR, Hartwig EE (1980) Resistance to soybean rust (Phakopsora pachyrhizi) and mode of inheritance. Crop Sci 20:254–255
Carter TE, Nelson RL, Sneller CH, Cui Z (2004) Genetic diversity in soybean. In: Boerma HR, Specht JE (eds) Soybeans: improvement, production and uses, 3rd edn. ASA-CSSA-SSSA, Madison, pp 303–416
Cho MH, Ciulla D, Klanderman BJ, Raby BA, Silverman EK (2008) High-resolution melting curve analysis of genomic and whole-genome amplified DNA. Clin Chem 54:2055–2058
Choi IY, Hyten DL, Matikumalli LK, Song Q, Chaky JM, Quigley CV, Chase K, Lark KG, Reiter RS, Yoon MS, Hwang EY, Yi SI, Young ND, Shoemaker RC, van Tassell CP, Specht JE, Cregan PB (2007) A soybean transcript map: gene distribution, haplotype and single nucleotide polymorphism analysis. Genetics 176:685–696
de Silva D, Blackett J (2007) Assay: high-resolution melting and unlabeled probes. Gen Eng Biotech News 27:3
Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucl Acids Res 19:1349
Garcia A, Calvo ES, Kiihl RAS, Harada A, Hiromoto DM, Vieira LGE (2008) Molecular mapping of soybean rust (Phakopsora pachyrhizi) resistance genes: discovery of a novel locus and alleles. Theor Appl Genet 117:545–553
Gizlice Z, Carter TE Jr, Burton JW (1993) Genetic diversity in North American soybean: I. Multivariate analysis of founding stock and relation to coefficient of parentage. Crop Sci 33:614–620
Gizlice Z, Carter TE Jr, Burton JW (1994) Genetic base for North American public soybean cultivars released between 1947 and 1988. Crop Sci 34:1143–1151
Grau CR, Dorrance AE, Bond J, Russin JS (2004) Fungal diseases. In: Boerma HR, Specht JE (eds) Soybeans: improvement, production and uses. Agron Monogr 16, 3rd edn. ASA-CSSA-SSSA, Madison, pp 679–763
GRIN (2009) Germplasm resources information network. Available at http://www.arsgrin.gov/npgs/index.html (verified 21 Dec. 2009). Natl Germplasm Resour Lab, Beltsville, MD
Ha BK, Boerma HR (2008) High-throughput SNP genotyping by melting curve analysis for resistance to southern root-knot nematode and frogeye leaf spot in soybean. J Crop Sci Biotech 11:91–100
Ha BK, Hussey RS, Boerma HR (2007) Development of SNP assays for marker-assisted selection of two Southern root-knot nematode resistance QTL in soybean. Plant Genome 2:S73–S82
Hartwig EE (1986) Identification of a fourth major gene conferring resistance to soybean rust. Crop Sci 26:1135–1136
Hartwig EE, Bromfield KR (1983) Relationships among three genes conferring specific resistance to rust in soybeans. Crop Sci 23:237–239
Hurley JD, Engle LJ, Davis JT, Welsh AM, Landers JE (2004) A simple, bead-based approach for multi-SNP molecular haplotyping. Nucleic Acids Res 32:e186
Hyten DL, Song Q, Zhu Y, Choi IY, Nelson RL, Costa JM, Specht JE, Shoemaker RC, Cregan PB (2006) Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci USA 102:16666–16671
Hyten DL, Hartman GL, Nelson RL, Frederick RD, Concibido VC, Narvel JM, Cregan PB (2007) Map location of the Rpp1 locus that confers resistance to soybean rust in soybean. Crop Sci 47:837–840
Hyten DL, Smith JR, Frederick RD, Tuchekr ML, Song Q, Cregan PB (2009) Bulked segregant analysis using the GoldenGate assay to locate the Rpp3 locus that confers resistance to soybean rust in soybean. Crop Sci 49:265–271
Keim P, Olson TC, Shoemaker RC (1988) A rapid protocol for isolating soybean DNA. Soy Genet Newslett 15:150–152
Kwok PY, Chen X (2003) Detection of single nucleotide polymorphisms. Curr Issues Mol Biol 5:43–60
Liew M, Pryor R, Palais R, Meadows C, Erali M, Lyon E, Wittwer C (2004) Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem 50:1156–1164
Liew M, Seipp M, Durtschi J, Margraf RL, Dames S, Erali M, Voelkerding K, Wittwer C (2007) Closed-tube SNP genotyping without labeled probes. Am J Clin Pathol 127:341–348
Margraf RL, Mao R, Wittwer CT (2008) Rapid diagnosis of MEN2B using unlabeled probe melting analysis and the LightCycler 480 instrument. J Mol Diagn 10:123–128
McLean RJ, Byth DE (1980) Inheritance of resistance to rust (Phakopsora pachyrhizi) in soybeans. Aust J Agric Res 31:951–956
Meyer JDF, Silva DCG, Yang C, Pedley KF, Zhang C, van de Mortel M, Hill JH, Shoemaker RC, Abdelnoor RV, Whitham SA, Graham MA (2009) Identification and analyses of candidate genes for Rpp4-mediated resistance to Asian soybean rust in soybean (Glycine max (L.) Merr.). Plant Physiol 150:295–307
Mohan M, Nair S, Bhagwat A, Krishna RG, Yano M, Bhatia CR, Sasaki T (1997) Genome mapping, molecular markers and marker-assisted selection in crop plants. Mol Breed 3:87–103
Monteros MJ, Missaoui AM, Phillips DV, Walker DR, Boerma HR (2007) Mapping and confirmation of the ‘Hyuuga’ red-brown lesion resistance gene for Asian soybean rust. Crop Sci 47:829–834
Pierozzi PHB, Ribeiro AS, Moreira JUV, Laperuta LDC, Rachid BF, Lima WF, Arias CAA, Oliveria MF, Toledo JFF (2008) New soybean (Glycine max Fabales, Fabaceae) sources of qualitative genetic resistance to Asian soybean rust caused by Phakospsora pachyrhizi (Uredinales, Phakopsoraceae). Genet Mol Biol 31:505–511
SantaLucia J, Allawi HT, Seneviratne PA (1996) Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry 35:3555–3562
Schneider RW, Hollier CA, Whitam HK, Palm ME, McKemy JM, Hernandez L, Levy L, De Vries-Paterson R (2005) First report of soybean rust caused by Phakopsora pachyrhizi in the continental United States. Plant Dis 89:774
Shastry BS (2004) Role of SNP/haplotype map in gene discovery and drug development: an overview. Drug Development Res 62:143–150
Shipe ER, Mueller JD, Lewis SA, Williams PF, Tomkins JP (1997) Registration of ‘Dillon’ soybean. Crop Sci 37:1983
Silva DCG, Yamanaka N, Brogin RL, Arias CAA, Nepomuceno AL, Di Mauro AO, Pereira SS, Nogueira LM, Passianotto ALL, Abdelnoor RV (2008) Molecular mapping of two loci that confer resistance to Asian rust in soybean. Theor Appl Genet 117:57–63
Sinclair JB, Hartman GL (1996) In: Proc soybean rust workshop, Urbana, I: Natl Soybean Res Lab Publ 1, Urbana-Champaign, IL
Singh BB, Thapliyal PN (1977) Breeding for resistance to soybean rust in India. In: Ford RE, Sinclair JB (eds) Rust of soybeans, the problem and research needs. Int Agric Publ INTSOY Ser. 12. University of Illinois, Urbana, IL, pp 62–65
Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Prior RJ (2003) High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem 49:853–860
Xu Y, Crouch JH (2008) Marker-assisted selection in plant breeding: from publications to practice. Crop Sci 48:391–407
Zhou L, Myers AN, Vandersteen JG, Wang L, Wittwer CT (2004) Closed-tube genotyping with unlabeled oligonucleotide probes and a saturating DNA dye. Clin Chem 50:1328–1335
Zhu YL, Song QJ, Hyten DL, Van Tassell CP, Matukumalli LK, Grimm DR, Hyatt SM, Fickus EW, Young ND, Cregan PB (2003) Single nucleotide polymorphisms in soybean. Genetics 163:1123–1134
Zhu S, Walker DR, Boerma HR, All JN, Parrott WA (2006) Fine mapping of a major insect resistance QTL in soybean and its interaction with minor resistance QTLs. Crop Sci 46:1094–1099
Acknowledgments
This research was supported by the Georgia Agricultural Experiment Stations and grants from the United Soybean Board. We would like to thank Randy Nelson (USDA-ARS, Urbana, IL) for providing the soybean seed of plant introductions containing the ASR resistance genes and the 32 ancestral lines and Perry Cregan and David Hyten (USDA-ARS, Beltsville, MD) for contributing the SNP primer sequences.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Xu.
Bo-Keun Ha contributed equally to this work.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Monteros, M.J., Ha, BK., Phillips, D.V. et al. SNP assay to detect the ‘Hyuuga’ red-brown lesion resistance gene for Asian soybean rust. Theor Appl Genet 121, 1023–1032 (2010). https://doi.org/10.1007/s00122-010-1368-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-010-1368-8