Abstract
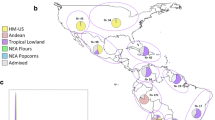
The highland region or Northwestern Argentina (NWA) is one of the southernmost areas of native maize cultivation and constitutes an expansion of the peruvian Andes sphere of influence. To examine the genetic diversity and racial affiliations of the landraces cultivated in this area, 18 microsatellite markers were used to characterize 147 individuals from 6 maize races representative of traditional materials. For the whole data set, a total of 184 alleles were found, with an average of 10.2 alleles per locus. The average gene diversity was 0.571. The observed patterns of genetic differentiation suggest that historical association is probably the main factor in shaping population structure for the landraces studied here. In agreement with morphological and cytogenetic data, Bayesian analysis of NWA landraces revealed the occurrence of three main gene pools. Assessment of racial affiliations using a combined dataset including previous data on American landraces showed a clear relationship between one of these gene pools and typical Andean races, whereas the remaining two gene pools exhibited a closer association to Caribbean accessions and native germplasm from the United States, respectively. These results highlight the importance of integrating regional genetic studies if a deeper understanding of maize diversification and dispersal is to be achieved.







Similar content being viewed by others
References
Anderson E, Cutler HC (1942) Races of Zea mays: I. Their recognition and classification. Ann Mo Bot Gard 29:69–89
Bracco M, Lia V, Gottlieb A, Cámara Hernández J, Poggio L (2009) Genetic diversity in maize landraces from indigenous settlements of Northeastern Argentina. Genetica 135:39–49
Bretting PK, Goodman MM, Stuber CW (1987) Karyological and isozyme variation in West Indian and allied American mainland races of maize. Am J Bot 74:1601–1613
Bretting PK, Goodman MM, Stuber CW (1990) Isozymatic variation in Guatemalan races of maize. Am J Bot 77:211–225
Brieger FG, Gurgel JTA, Paterniani E, Blumenschein A, Alleoni MR (1958) The races of maize in Brazil and other eastern South American countries. National Academy of ScienceNational Research Council Publications, Washington, DC
Brunson AM, Bower CW (1931) Pop corn. USDA Farmers′ Bull 1679:17
Brush SB, Perales H (2007) A maize landscape: ethnicity and agro-biodiversity in Chiapas Mexico. Agric Ecosyst Environ 121:211–221
Cámara Hernández J, Miante Alzogaray AM (1997) Las razas de maíz de Jujuy y Salta Argentina. Proc 1er Taller Internacional de Recursos Fitogenéticos del Noroeste Argentino 1:19–23
Camus-Kulandaivelu L, Veyrieras JB, Madur D et al (2006) Maize adaptation to temperate climate: relationship between population structure and polymorphism in the Dwarf8 gene. Genetics 172(4):2449–2463
Camus-Kulandaivelu L, Veyrieras J-B, Gouesnard B, Charcosset A, Manicacci D (2007) Evaluating the reliability of structure outputs in case of relatedness between individuals. Crop Sci 47:887–890
Colasanti J,ZY, Sundaresan V (1998) The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93:593–603
Doebley J, Goodman MM (1984) Isoenzymatic variation in Zea (Gramineae). Syst Bot 9:203–218
Doebley J, Goodman MM, Stuber CW (1983) Isozyme variation in maize from the southwestern United States taxonomic and anthropological implications. Maydica 28:94–120
Doebley J, Goodman MM, Stuber CW (1985) Isozyme variation in the races of maize from Mexico. Am J Bot 72:629–639
Doebley J, Goodman MM, Stuber CW (1986) Exceptional genetic divergence of Northern Flint Corn. Am J Bot 73:64–69
Doebley J, Goodman MM, Stuber CW (1987) Patterns of isozyme variation between maize and Mexican Annual teosinte. Econ Bot 41:234–246
Doebley J, Wendel JF, Smith JSC, Stuber CW, Goodman MM (1988) The origin of cornbelt maize: the isozyme evidence. Econ Bot 42:120–131
Dubreuil P, Charcosset A (1998) Genetic diversity within and among maize populations: a comparison between isozyme and nuclear RFLP loci. Theor Appl Genet 96:577–587
El Mousadik A, Petit R (1996) High level of genetic differentiation for allelic richness among populations of the argan tree (Argania spinosa (L.) Skeels) endemic to Morocco. Theor Appl Genet 92:832–839
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure: extensions to linked loci and correlated allele frequencies. Genetics 164:1567–1587
Felsenstein J (1991–2004) Phylip (Phylogeny Inference Package). University of Washington, Washington
Gaggiotti OE, Lange O, Rassmann K, Gliddon C (1999) A comparison of two indirect methods for estimating average levels of gene flow using microsatellite data. Mol Ecol 8:1513–1520
Gale MD, Devos KM (1998) Comparative genetics in the grasses. Proc Natl Acad Sci USA 95:1971–1974
Goodman MM (1990) Genetic and germplasm stocks worth conserving. J Hered 81:11–16
Goodman MM (2005) Broadening the U.S. germplasm base. Maydica 50:203–214
Goodman MM, Bird R McK (1977) The races of Maize IV: tentative grouping of 219 Latin American Races. Econ Bot 31:204–221
Goodman MM, Brown WL (1988) Races of corn. In: Sprague GF, Dudley JW (eds) Corn and corn improvement. Amer Soc Agron, Madison, WI, pp 33–79
Goodman MM, Stuber CW (1983) Races of maize. VI. Isozyme variation among races of maize in Bolivia. Maydica 28:169–187
Goudet J (1995) Fstat version 1.2: a computer program to calculate F-statistics. J Hered 86:485–486
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available from http://www.unil.ch/izea/softwares/fstat.html. Updated from Goudet 1995
Hardy OJ, Charbonnel N, Freville H, Heuertz M (2003) Microsatellite allele sizes: a simple test to assess their significance on genetic differentiation. Genetics 163:1467–1482
Hoisington D, Khairallah M, Reeves T et al (1999) Plant genetic resources: what can they contribute toward increased crop productivity? Proc Natl Acad Sci USA 96:5937–5943
Horovitz S (1935) Distribución geográfica de factores genéticos en los maíces autóctonos del norte argentino. Rev Arg Agron 2:133–135
Kahler AL, Hallauer AR, Gardner C (1986) Allozyme polymorphism within and among open-pollinated and adapted exotic populations of maize. Theor Appl Genet 72:592–601
Lia VV, Confalonieri VA, Poggio L (2007) B chromosome polymorphism in maize landraces: adaptive vs. demographic hypothesis of clinal variation. Genetics 177:895–904
Liu K, Goodman M, Muse S et al (2003) Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics 165:2117–2128
Matsuoka Y, Vigouroux Y, Goodman MM et al (2002a) A single domestication for maize shown by multilocus microsatellite genotyping. Proc Natl Acad Sci USA 99:6080–6084
Matsuoka Y, Mitchell SE, Kresovich S, Goodman MM, Doebley JF (2002b) Microsatellites in Zea—variability, patterns of mutations, and use for evolutionary studies. Theor Appl Genet 104:436–450
McClintock B, Kato TA, Blumenschein A (1981) Chromosome constitution of the races of maize, its significance in the interpretation of relationships between races and varieties in the Americas. Colegio de Postgraduados, Chapingo
Minch E, Ruiz Linares A, Goldstein D, Feldman M, Cavalli-Sforza LL (1996) Microsat (version 1.5b): a computer program for calculating various statistics on microsatellite allele data. (Program available from: http://hpgl.stanford.edu/projects/microsat/)
Nei M (1972) Genetic distance between populations. Am Nat 106:283–292
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nielsen R, Slatkin M (2000) Likelihood analysis of ongoing gene flow and historical association. Evolution 54:44–50
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155
Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Perales RH, Benz BF, Brush SB (2005) Maize diversity and ethnolinguistic diversity in Chiapas, Mexico. Proc Natl Acad Sci USA 102:949–954
Poggio L, Rosato M, Chiavarino AM, Naranjo CA (1998) Genome size and environmental correlations in maize (Zea mays ssp. mays, Poaceae). Ann Bot (Lond) 82:107–115
Pressoir G, Berthaud J (2004) Patterns of population structure in maize landraces from the Central Valleys of Oaxaca in Mexico. Heredity 92:88–94
Pritchard JK, Wen W (2003) Documentation for STRUCTURE software: Version 2. http://pritch.bsd.uchicago.edu
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Reif JC, Xia XC, Melchinger AE et al (2004) Genetic diversity determined within and among CIMMYT maize populations of tropical, subtropical, and temperate germplasm by SSR markers. Crop Sci 44:326–334
Reif J, Warburton M, Xia X et al (2006) Grouping of accessions of Mexican races of maize revisited with SSR markers. Theor Appl Genet 113:177–185
Rice W (1989) Analysing tables of statistical tests. Evolution 43:223–225
Roberts LM, Grant UJ, Ramírez ER, Hatheway WH, PLwPCM Smith (1957) Races of Maize in Colombia. National Academy of Science–National Research Council Publications, Washington, DC
Rosato M, Chiavarino AM, Naranjo CA, Cámara Hernandez J, Poggio L (1998) Genome size and numerical polymorphism for the B chromosome in races of maize (Zea mays ssp. mays, Poaceae). Am J Bot 85:168–174
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sanchez GJ, Stuber CW, Goodman MM (2000) Isozymatic diversity in the races of maize of the Americas. Maydica 45:185–203
Santacruz-Varela A, Widrlechner MP, Ziegler KE et al (2004) Phylogenetic relationships among North American popcorns and their evolutionary links to Mexican and South American popcorns. Crop Sci 44:1456
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462
Taller JM, Bernardo R (2004) Diverse adapted populations for improving northern maize inbreds. Crop Sci 44:1444–1449
Tarter JA, Goodman MM, Holland JB (2003) Testcross performance of semiexotic inbred lines derived from Latin American maize accessions. Crop Sci 43:2272–2278
Thornsberry JM, Goodman MM, Doebley J et al (2001) Dwarf8 polymorphisms associate with variation in flowering time. Nat Genet 28:203–204
Torregrosa M, Cámara Hernandez J, Solari L et al (1980) Clasificación preliminar de formas raciales de maíz y su distribución geográfica en la República Argentina. Actas II Congreso Nacional de Maíz 1:5–17
Vigouroux Y, Glaubitz JC, Matsuoka Y et al (2008) Population structure and genetic diversity of New World maize races assessed by DNA microsatellites. Am J Bot 95:1240–1253
Waples RS, Gaggiotti O (2006) What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol Ecol 15:1419–1439
Warburton ML, Xianchua X, Crossa J et al (2002) Genetic characterization of CIMMYT inbred lines and open pollinated populations using large scale fingerprinting methods. Crop Sci 42:1832–1840
Weir B, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Wellhausen EJ, Roberts LM, Hernandez EMP (1952) Races of maize in Mexico. Harvard University Press, Cambridge
Wright S (1978) Evolution and the genetics of populations, volume 4: variability within and among natural populations, 3rd edn. University of Chicago Press, Chicago
Acknowledgments
The authors wish to thank Ing. J. Cámara Hernández and A.M. Miante Alzogaray for helpfull discussion on racial characteristics. We are grateful to Dr. J. Doebley and Dr. Y. Matsuoka for kindly providing us the SSR data from Matsuoka et al. (2002a) and Liu et al. (2003). We are also in debt to Dr. Jim Holland, for generously sending us the inbred lines used to establish allele equivalencies between datasets; and to the anonymous reviewers who have greatly improved the manuscript. Several grants from the Consejo Nacional de Investigaciones Científicas y Técnicas (PIP 2296, PIP 5927), the Universidad de Buenos Aires (EX-317), and the Agencia Nacional de Promoción Científica y Tecnológica (BID 1201 OC-AR PICT 04443, BID 1728 OC-AR583 PICT 14119) are gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Charcosset.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2009_1108_MOESM1_ESM.pdf
Fig. S1 Neighbor-joining tree of 71 STRUCTURE outputs derived from the analysis of population structure in maize landraces from Northwestern Argentina. Distances among outputs were calculated following the methods of Camus-Kulandaivelu et al. (2007) (PDF 14 kb)
122_2009_1108_MOESM2_ESM.pdf
Fig. S2 Neighbor-joining tree of 71 STRUCTURE outputs derived from the analysis of population structure in a combined dataset including maize landraces from Northwestern Argentina and a set of American landraces from Matsuoka et al. (2002a). Distances among outputs were calculated following the methods of Camus-Kulandaivelu et al. (2007) (PDF 14 kb)
Rights and permissions
About this article
Cite this article
Lia, V.V., Poggio, L. & Confalonieri, V.A. Microsatellite variation in maize landraces from Northwestern Argentina: genetic diversity, population structure and racial affiliations. Theor Appl Genet 119, 1053–1067 (2009). https://doi.org/10.1007/s00122-009-1108-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-009-1108-0