Abstract
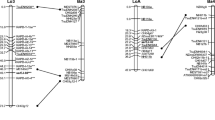
Tef [Eragrostis tef (Zucc.) Trotter] is the major cereal crop in Ethiopia. Tef is an allotetraploid with a base chromosome number of 10 (2n = 4x = 40) and a genome size of 730 Mbp. Ninety-four F9 recombinant inbred lines (RIL) derived from the interspecific cross, Eragrostis tef cv. Kaye Murri × Eragrostis pilosa (accession 30-5), were mapped using restriction fragment length polymorphisms (RFLP), simple sequence repeats derived from expressed sequence tags (EST–SSR), single nucleotide polymorphism/insertion and deletion (SNP/INDEL), intron fragment length polymorphism (IFLP) and inter-simple sequence repeat amplification (ISSR). A total of 156 loci from 121 markers was grouped into 21 linkage groups at LOD 4, and the map covered 2,081.5 cM with a mean density of 12.3 cM per locus. Three putative homoeologous groups were identified based on multi-locus markers. Sixteen percent of the loci deviated from normal segregation with a predominance of E. tef alleles, and a majority of the distorted loci were clustered on three linkage groups. This map will be useful for further genetic studies in tef including mapping of loci controlling quantitative traits (QTL), and comparative analysis with other cereal crops.


Similar content being viewed by others
References
Ayele M, Dolezel J, Van Duren M, Brunner H, Zapata-Arias FJ (1996) Flow cytometric analysis of nuclear genome of the Ethiopian cereal tef [Eragrostis tef (Zucc.) Trotter]. Genetics 98:211–215
Bai G, Tefera H, Ayele M, Nguyen HT (1999) A genetic linkage map of tef [Eragrostis tef (Zucc.) Trotter] based on amplified fragment length polymorphism. Theor Appl Genet 99:599–604
Barrett B, Griffiths A, Schreiber M, Ellison N, Mercer C, Bouton J, Ong B, Forster J, Sawbridge T, Spangenberg G, Bryan G, Woodfield D (2004) A microsatellite map of white clover. Theor Appl Genet 109:596–608
Blanco A, Bellomo MP, Cenci A, Giovanni CD, D’Ovidio R, Iacono E, Laddomada B, Pagnotta MA, Porceddu E, Sciancalepore A, Simeone R, Tanzarella OA (1998) A genetic linkage map of durum wheat. Theor Appl Genet 97:721–728
Causse MA, Fulton TM, Cho YG, Ahn SN, Chunwongse J, Wu K, Xiao J, Yu X, Ronald PC, Harrington SE, Second G, McCouch SR, Tanksley SD (1994) Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics 138:1251–1274
Central Statistical Authority (2004) Statistical abstract of Ethiopia 2004 (1996 EC). Addis Ababa, Ethiopia
Chakravarti A, Lasher LK, Reefer JE (1991) A maximum likelihood method for estimating genome length using genetic linkage data. Genetics 128:175–182
Devos KM (2005) Updating the ‘crop circle’. Curr Opin Plant Biol 8:155–162
Duran Y, Fratini R, Garcia P, Perez de la Vega M (2004) An intersubspecific genetic map of Lens. Theor Appl Genet 108:1265–1273
Faris JD, Laddomada B, Gill BS (1998) Molecular mapping of segregation distortion loci in Aegilops tauschii. Genetics 149:319–327
Feltus FA, Singh HP, Lohithaswa HC, Schulze SR, Silva TD, Paterson AH (2006) A comparative genomics strategy for targeted discovery of single-nucleotide polymorphisms and conserved-noncoding sequences in orphan crops. Plant Physiol 140:1183–1191
Fishman L, Kelly AJ, Morgan E, Willis JH (2001) A genetic map in the Mimulus guttatus species complex reveals transmission ratio distortion due to heterospecific interactions. Genetics 159:1701–1716
Gale MD, Devos KM (1998) Comparative genetics in the grasses. Proc Natl Acad Sci USA 95:1971–1974
Gardiner GM, Melia-Hancock S, Hoisington DA, Chao S (1993) Development of a core RFLP map in maize using an immortalized F2 population. Genetics 134:917–930
Han Z, Wang C, Song X, Guo W, Gou J, Li C, Chen X, Zhang T (2006) Characteristics, development and mapping of Gossypium hirsutum derived EST–SSRs in allotetraploid cotton. Theor Appl Genet 112:430–439
Heun M, Kennedy AE, Anderson JA, Lapitan NL, Sorrells ME, Tanksley SD (1991) Construction of a restriction fragment length polymorphism map for barley (Hordeum vulgare). Genome 34:437–447
Holloway JL, Knapp SJ (1993) Gmendel 3.0 users guide. Department of Crop and Soil Science, Oregon State University, OR
Hu J, Vick BA (2003) Targeted region amplification polymorphism: a novel marker technique for plant genotyping. Plant Mol Biol Rep 21:289–294
Kantety RV, Zeng X, Bennetzen JL, Zehr BE (1995) Assessment of genetic diversity in dent and popcorn (Zea mays L.) inbred lines using inter-simple sequence repeat (ISSR) amplification. Mol Breed 1:365–373
Ketema S (1993) Tef (Eragrostis tef): breeding, genetic resources, agronomy, utilization and role in Ethiopian agriculture. Institute of Agricultural Research, Addis Ababa, Ethiopia
Korzun V, Roder MS, Wendehake K, Pasqualone A, Lotti C, Ganal MW, Blanco A (1999) Integration of dinucleotide microsatellites from hexaploid bread wheat into a genetic linkage map of durum wheat. Theor Appl Genet 98:1202–1207
Kosambi DD (1944) The estimation of map distance from recombination values. Ann Eugen 12:172–175
Lange K, Boehnke M (1982) How many polymorphic genes will it take to span the human genome? Am J Hum Genet 34:842–845
Lyttle TW (1991) Segregation distorters. Annu Rev Genet 25:511–557
Martin-Lopes P, Zhang H, Koebner R (2001) Detection of single nucleotide mutations in wheat using single strand conformation polymorphism gels. Plant Mol Biol Rep 19:159–162
Manly KF, Olson JM (1999) Overview of QTL mapping software and introduction to Map Manager QT. Mamm Genome 10:327–334
Nelson JC, Van Deynze AE, Autrique E, Sorrells ME, Liu YH, Merlino M, Atkinson M, Leroy P (1995a) Molecular mapping of wheat: homeologous group 2. Genome 38:517–524
Nelson JC, Van Deynze AE, Autrique E, Sorrells ME, Liu YH, Negre S, Bernard M, Leroy P (1995b) Molecular mapping of wheat: homeologous group 3. Genome 38:525–533
Nelson JC, Sorrells ME, Van Deynze AE, Liu YH, Autrique E, Atkinson M, Bernard M, Leroy P (1995c) Molecular mapping of wheat: major genes and rearrangements in homeologous groups 4, 5 and 7. Genetics 141:721–731
O’Donoughue LS, Kinian SF, Rayapati PJ, Panner GA, Sorrells M, Tanksley SD, Phillips RL, Rines HW, Lee M, Fedak G, Molnar SJ, Hoffman D, Salas CA, Wu B, Autrique E, Van Deynze A (1995) A molecular linkage map of cultivated oat. Genome 38:368–380
Sorrells ME (2001) Comparative genomics for tef improvement. In: Tefera H, Belay G, Sorrells M (eds). Narrowing the rift: tef research and development. Proceedings of the international workshop on tef genetics and improvement, 16–19 October 2000, Addis Ababa, Ethiopia, pp 109–119
Temnykh S, Park WD, Ayers N, Cartinhour S, Hauck N, Lipovich L, Cho YG, Ishii T, McCouch SR (2000) Mapping and genome organization of microsatellites in rice (Oryza sativa). Theor Appl Genet 100:698–712
Van Deynze AE, Dubcovsky J, Gill KS, Nelson JC, Sorrells ME, Dvorak J, Gill BS, Lagudah ES, McCouch SR, Appels R (1995) Molecular-genetic maps for group 1 chromosomes of Triticeae species and their relation to chromosomes in rice and oat. Genome 38:45–59
Wendel JF (2000) Genome evolution in polyploids. Plant Mol Biol 42:225–249
Xu Y, Zhu L, Xiao J, Huang N, McCouch SR (1997) Chromosomal regions associated with segregation distortion of molecular markers in F2, backcross, doubled haploid, and recombinant inbred populations in rice (Oryza sativa L.). Mol Gen Genet 253:535–545
Yu J-K, Dake TM, Singh S, Benscher D, Li W, Gill B, Sorrells ME (2004a) Development and mapping of EST-derived simple sequence repeat markers for hexaploid wheat. Genome 47:805–818
Yu J-K, La Rota M, Kantety RV, Sorrells ME (2004b) EST derived SSR markers for comparative mapping in wheat and rice. Mol Genet Genomics 271:742–751
Yu J-K, Sun Q, La Rota M, Edwards H, Tefera H, Sorrells ME (2006) Expressed sequence tag analysis in tef [Eragrostis tef (Zucc.) Trotter]. Genome 49:365–372
Zhang D, Ayele M, Tefera H, Nguyen HT (2001) RFLP linkage map of the Ethiopian cereal tef [Eragrostis tef (Zucc.) Trotter]. Theor Appl Genet 102:957–964
Acknowledgments
J.-K. Yu and R.V. Kantety contributed equally to the research described herein. The authors would like to acknowledge financial support from the McKnight Foundation Collaborative Crop Research Program (Grant # 00-933) and Hatch project 149419. We are grateful to Drs. Susan McCouch at Cornell University, Bikram Gill at Kansas State University, Andrew Paterson and Katrien Devos at University of Georgia for providing the primers of RM, KSUM, SRSC/PRSC and inf/lfm markers, respectively. We also wish to acknowledge R. Greene, C. Bermudez and C. T. Jayasuriya for their generous assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Gebhardt.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Yu, JK., Kantety, R.V., Graznak, E. et al. A genetic linkage map for tef [Eragrostis tef (Zucc.) Trotter]. Theor Appl Genet 113, 1093–1102 (2006). https://doi.org/10.1007/s00122-006-0369-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-006-0369-0