Abstract
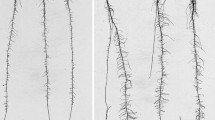
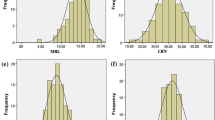
Brassica oleracea can be genetically engineered using Agrobacterium rhizogenes. The initial stage of this process is the production of transgenic (‘hairy’) roots; shoots are subsequently regenerated from these roots. Previous work using gus and gfp reporter genes has shown that genotypes of B. oleracea vary in their performance for transgenic root production. Quantitative trait loci (QTLs) controlling this trait have been located in one mapping population. The current study provides evidence that performance for transgenic root production is associated with performance for adventitious (non-transgenic) root production in B. oleracea across a second mapping population. This is shown by regression analyses between performance for the two traits and the demonstration that QTLs controlling the two traits map to the same positions within the genome. Since the rate of adventitious root production does not differ significantly in the presence and absence of A. rhizogenes, there is no evidence that the expression of Agrobacterium genes induces adventitious root production. It is apparent that genotypes exhibiting high adventitious root production in the absence of A. rhizogenes will also tend to show high transgenic root production, thereby allowing the selection of lines that are more efficiently transformed.


Similar content being viewed by others
References
Ballas N, Citovsky V (1997) Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of Agrobacterium VirD2 protein. Proc Natl Acad Sci USA 94:10723–10728
Barg R, Pilowsky M, Shabtai S, Carmi N, Szechtman AD, Dedicova B, Salts Y (1997) The TYLCV-tolerant tomato line MP-1 is characterized by superior transformation competence. J Exp Bot 48:1919–1923
Chateau S, Sangwan RS, Sangwan-Norreel BS (2000) Competence of Arabidopsis thaliana genotypes and mutants for Agrobacterium tumefaciens-mediated gene transfer: role of phytohormones. J Exp Bot 51:1961–1968
Cogan N, Harvey E, Robinson H, Lynn J, Pink D, Newbury HJ, Puddephat I (2001) The effects of anther culture and plant genetic background on Agrobacterium rhizogenes-mediated transformation of commercial cultivars and derived doubled haploid Brassica oleracea. Plant Cell Rep 20:755–762
Cogan NOI, Lynn JR, King GJ, Kearsey MJ, Newbury HJ, Puddephat IJ (2002) Identification of genetic factors controlling the efficiency of Agrobacterium rhizogenes-mediated transformation of Brassica oleracea by QTL analysis. Theor Appl Genet 105:568–576
Cogan NOI, Newbury HJ, Oldacres AM, Lynn JR, King GJ, Kearsey MJ, Puddephat IJ (2004) Identification and characterisation of QTL controlling Agrobacterium-mediated transient and stable transformation of Brassica oleracea. Plant Biotechnol 2:59–69
Costacurta A, Vanderleyden J (1995) Synthesis of phytohormones by plant-associated bacteria. Crit Rev Microbiol 21:1–18
Deng W, Chen LS, Wood DW, Metcalfe T, Liang XY, Gordon MP, Comai L, Nester EW (1998) Agrobacterium VirD2 protein interacts with plant host cyclophilins. Proc Natl Acad Sci USA 95:7040–7045
Falasca G, Reverberi M, Lauri P, Caboni E, De Stradis A, Altamura NM (2000) How Agrobacterium rhizogenes triggers de novo root formation in a recalcitrant woody plant: an integrated histological, ultrastructural and molecular analysis. New Phytol 145:77–93
Gelvin SB (2000) Agrobacterium and plant genes involved in T-DNA transfer and integration. Annu Rev Plant Physiol Plant Mol Biol 51:223–256
Gelvin SB (2003) Improving plant genetic engineering by manipulating the host. Trends Biotechnol 21:95–98
Gelvin SB, Liu CN (1994) Genetic manipulation of Agrobacterium tumefaciens strains to improve transformation of recalcitrant plant species. In: Gelvin SB, Schilperoort RA (eds) Plant molecular biology manual. Kluwer, Dordrecht, B4, p 1013
Guerche P, Jouanin L, Tepfer D, Pelletier G (1987) Genetic transformation of oilseed rape (Brassica napus) by the Ri T-DNA of Agrobacterium rhizogenes and analysis of the inheritance of the transformed phenotype. Mol Gen Genet 206:382–386
Haley CS, Knott SA (1992) A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69:315–324
Haseloff J, Siemering KR, Prasher DC, Hodge S (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94:2122–2127
Jansen RC, Stam P (1994) High resolution of quantitative traits into multiple loci via interval mapping. Genetics 136:1447–1455
Kearsey MJ, Hyne V (1994) QTL analysis—a simple marker regression approach. Theor Appl Genet 89:698–702
Kearsey MJ, Pooni HS (1998) The genetic analysis of quantitative traits. Chapman and Hall, London
Lander ES, Botstein D (1989) Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121:185–199
McCullaugh P, Nelder JA (1987) Generalized linear models. Chapman and Hall, London
Morris RO, Blevins DG, Dietrich JT, Durley RC, Gelvin SB, Gray J, Hommes NG, Kaminek M, Mathews LJ, Meilan R, Reinbott TM, Sayavedrasoto L (1993) Cytokinins in plant pathogenic bacteria and developing cereal grains. Aust J Plant Physiol 20:621–637
Murashige T, Skoog FS (1962) A revised method for rapid growth and bioassays and tobacco tissue culture. Physiol Plant 15:437–497
Mysore KS, Nam J, Gelvin SB (2000) An Arabidopsis histone H2A mutant is deficient in Agrobacterium T-DNA integration. Proc Natl Acad Sci USA 97:948–953
Nam J, Mattysse AG, Gelvin SB (1997) Differences in susceptibility of Arabidopsis ecotypes to crown gall disease may result from a deficiency in T-DNA integration. Plant Cell 9:317–333
Nam J, Mysore KS, Zheng C, Knue MK, Mattysse AG, Gelvin SB (1999) Identification of T-DNA tagged Arabidopsis mutants that are resistant to transformation by Agrobacterium. Mol Gen Genet 261:429–438
Poulsen GB (1996) Genetic transformation of Brassica. Plant Breed 115:209–225
Puddephat IJ (2003) Plant genetic engineering. In: Newbury HJ (ed) Plant molecular breeding. Blackwell, Oxford, pp 82–133
Puddephat IJ, Riggs TJ, Fenning TM (1996) Transformation of Brassica oleracea L: a critical review. Mol Breed 2:185–210
Puddephat IJ, Robinson HT, Fenning TM, Barbara DJ, Morton A, Pink DAC (2001) Recovery of phenotypically normal transgenic plants of Brassica oleracea upon Agrobacterium rhizogenes-mediated co-transformation and selection of transformed hairy roots by GUS assay. Mol Breed 7:229–242
Rae A, Howell E, Kearsey MJ (1999) More QTL for flowering time revealed by substitution lines in Brassica oleracea. Heredity 83:586–596
Ramsey LD, Jennings DE, Bohoun EJR, Arthur AE, Lydiate DJ, Kearsey MJ, Marshall DF (1996) The construction of a substitution library of recombinant backcross lines in Brassica oleracea for the precise mapping of quantitative trait loci. Genome 39:558–567
Sangwan RSY, Bourgeois Y, Sangwannorreel BS (1991) Genetic transformation of Arabidopsis thaliana zygotic embryos and identification of critical parameters influencing transformation efficiency. Mol Gen Genet 230:475–485
Sebastian RL, Howell EC, King GJ, Marshall DF, Kearsey MJ (2000) An integrated AFLP and RFLP Brassica oleracea linkage map from two morphologically distinct doubled-haploid mapping populations. Theor Appl Genet 100:75–81
Spano L, Pomponi M, Constantino P, van Slogteren GMS, Tempe J (1982) Identification of T-DNA in the root-inducing plasmid of the agropine type Agrobacterium rhizogenes 1855. Plant Mol Biol 1:291–300
Sparrow PAC, Townsend TM, Arthur AE, Dale PJ, Irwin JA (2004a) Genetic analysis of Agrobacterium tumefaciens susceptibility in Brassica oleracea. Theor Appl Genet 108:644–650
Sparrow PAC, Townsend TM, Morgan CL, Dale PJ, Arthur AE, Irwin JA (2004b) Genetic analysis of in vitro shoot regeneration from cotyledonary petioles of Brassica oleracea. Theor Appl Genet 108:1249–1255
Sparrow PAC, Dale PJ, Irwin JA (2004c) The use of phenotypic markers to identify Brassica oleracea genotypes for routine high-throughput Agrobacterium-mediated transformation. Plant Cell Rep 23:64–70
Swart S, Lugtenberg BJJ, Smit G, Kijne JW (1994) Purification and partial characterisation of a glycoprotein from pea (Pisum sativum) with receptor activity for rhicadhesin, an attachment protein of the Rhizobiaceae. Plant Mol Biol 24:171–183
Tepfer D (1990) Genetic transformation using Agrobacterium rhizogenes. Physiol Plant 79:140–146
Tzfira T, Citovsky V (2000) From host recognition to T-DNA integration: the function of bacterial and plant genes in the Agrobacterium-plant cell interaction. Mol Plant Pathol 1:201–212
Tzfira T, Citovsky V (2002) Partners-in-infection: host proteins involved in the transformation of plant cells by Agrobacterium. Trends Cell Biol 12:121–129
Tzfira T, Vaidya M, Citovsky V (2001) VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J 20:3596–3607
Wagner VT, Mattysse AG (1992) Involvement of vitronectin-like protein in attachment of Agrobacterium tumefaciens to carrot suspension cells. J Bacteriol 174:5999–6003
Walkerpeach CR, Velten J (1994) Agrobacterium-mediated gene transfer to plant cells; cointegrate and binary vector systems. In: Gelvin SB, Schilperoort RA (eds) Plant molecular biology manual. Kluwer, Dordrecht, B4, pp 1–9
Wordragen MF van, Ouwerkerk PBF, Dons HJM (1992) Agrobacterium rhizogenes-mediated induction of apparently untransformed roots and callus in chrysanthemum. Plant Cell Tissue Organ Cult 30:149–157
Yibrah HS, Gronroos R, Lindroth A, Franzen H, Clapham D, von Arnold S (1996) Agrobacterium rhizogenes mediated induction of adventitious rooting from Pinus contorta hypocotyls and the effect of 5-azacytidine on transgene activity. Transgenic Res 5:75–85
Zhang J, Boone L, Kocz R, Zhang C, Binns A, Lynn DG (2000) At the maize/Agrobacterium interface: natural factors limiting host transformation. Chem Biol 7:611–621
Ziemienowicz A, Tinland B, Bryant J, Gloeckler V, Hohn B (2000) Plant enzymes but not Agrobacterium VirD2 mediate T-DNA ligation in vitro. Mol Cell Biol 20:6317–6322
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by G. Wenzel
Rights and permissions
About this article
Cite this article
Oldacres, A.M., Newbury, H.J. & Puddephat, I.J. QTLs controlling the production of transgenic and adventitious roots in Brassica oleracea following treatment with Agrobacterium rhizogenes. Theor Appl Genet 111, 479–488 (2005). https://doi.org/10.1007/s00122-005-2037-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-005-2037-1