Abstract
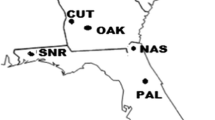
Rooted cuttings of 40 different clones (genotypes) of Picea abies (L.) Karst were planted in seven test environments (P, H, L, S, B, M and K) in northern Germany. Type-B genetic correlations between the pairs of test sites were estimated, and a cluster diagram based on inter-site genetic correlations for height was used to illustrate the general pattern of similarities among the test sites. Principal component analysis (PCA) showed that the first three principal components explained 97% of the variance in height, thereby providing a good summary of the relationships among the test sites. Test sites B and K accounted for a relatively high—and test sites H and M for a relatively low—proportion of genotype-environment interaction sums of squares and, consequently, for “ecovalence”. “Moderator” and “instigator” test sites for interactions in different site combinations were detected. On the basis of several similarity measures, we observed two distinct groups: sites M-S-P formed one group, while sites K-L formed the other; sites H and B were closer to the first group. The empirical data indicated that a good test site should have the following features: (1) low interaction behaviour, (2) low coefficient of variation, (3) high genotypic selectivity, (4) high coefficient of determination, (5) high efficiency of expected gain. Based on these criteria, site M was determined to be the best test site for screening genotypes to be planted in other environments. We concluded that one broadly adapted vegetative propagation population of P. abies can be used for the potential planting environments in northern Germany.



Similar content being viewed by others
References
Adams WT, White TL, Hodge GR, Powell GL (1994) Genetic parameters for bole volume in longleaf pine—large sample estimates and influences of test characteristics. Silvae Genet 43:357–366
Becker HC, Léon J (1988) Stability analysis in plant breeding. Plant Breed 101:1–23
Burdon RD (1977) Genetic correlation as a concept for studying genotype—environment interaction in forest tree breeding. Silvae Genet 26:168–175
Dieters MJ, White TL, Hodge GR (1995) Genetic parameter estimates for volume from full-sib tests of slash pine (Pinus elliottii). Can J For Res 25:1397–1408
Falconer DS (1952) The problem of environment and selection. Am Nat 86:293–298
Falconer DS (1981) Introduction to quantitative genetics, 2nd edn. Longman, London, New York
Finlay KW, Wilkinson GN (1963) The analyses of adaptation in a plant breeding programme. Aust J Agric Res 14:742–754
Gwaze DP, Bridgewater FE, Byram TD, Lowe WJ (2001) Genetic parameter estimates for growth and wood density in loblolly pine (Pinus taeda L.). For Genet 8(1):47–55
Hodge GR, White TL (1992) Genetic parameter estimates for growth traits at different ages in slash pine and some implications for breeding. Silvae Genet 41:252–262
Isik K, Kleinschmit J (2003) Stability related parameters and their evaluation in a 17-year old Norway spruce clonal test series. Silvae Genet 52:133–138
Isik K, Kleinschmit J, Svolba J (1995) Survival, growth trends and genetic gains in 17-year old Picea abies clones at seven test sites. Silvae Genet 44:116–128
Johnson GR (1997) Site-to-site genetic correlations and their implications on breeding zone size and optimum number of progeny test sites for coastal Douglas-fir. Silvae Genet 46:280–285
Johnson RA, Wichern DW (2002) Applied multivariate statistical analysis, 5th edn. Prentice Hall, New York
Karlsson B, Wellendorf H, Roulund H, Werner M (2001) Genotype × trial interaction and stability across sites in 11 combined provenance and clone experiments with Picea abies in Denmark and Sweden. Can J For Res 31:1826–1836
Kleinschmit J (1974) A program of large-scale cutting propagation of Norway spruce. N Z J For Sci 4:359–366
Koots KR, Gibson JB (1996) Realized sampling variances of estimates of genetic parameters and the difference between genetic and phenotypic correlations. Genetics 143:1409–1416
Lambeth CC, van Biujtenen JP, Duke SD (1983) Early selection is effective in 20-year-old genetic tests of loblolly pine. Silvae Genet 32:210–215
Li B, McKeand SE (1989) Stability of loblolly pine families in the southeastern US. Silvae Genet 38:96–101
Lindgren D (1993) The population biology of clonal deployment. In: Ahuja MR, Libby WJ (eds) Clonal forestry I. Genetics and biotechnology. Springer, Berlin Heidelberg New York, pp 34–49
Lu P, White TL, Huber DA (1999) Estimating type B genetic correlations with unbalanced data and heterogeneous variances for half-sib experiments. For Sci 45:562–572
Lu P, Huber DA, White TL (2001) Comparison of multivariate and univariate methods for the estimation of type B genetic correlations. Silvae Genet 50:13–22
McKeand SE, Li B, Hatcher AV, Weir RJ (1990) Stability parameter estimates for stem volume for loblolly pine families growing in different regions in the southeastern United States. For Sci 26:10–17
Nassar R, Léon J, Huehn M (1994) Tests of significance for combined measures of plant stability and performance. Biometric J 36:109–123
Owino F (1977) Genotype-environment interaction and genotypic stability in loblolly pine: II Genotypic stability comparisons. Silvae Genet 26:21–25
Riemenschnider DE (1988) Heritability, age-age correlations, and inferences regarding juvenile selection in jack pine. For Sci 34:1076–1082
SAS Institute (1987) SAS/STAT guide for personal computers, 6th edn. Cary, N.C.
Shelbourne CJA (1972) Genotype-environment interaction: its study and its implications in forest tree improvement. In: IUFRO Genet-SABRAO Joint Symp, Tokyo, p 27
Sneath PHA, Sokal RR (1973) Numerical taxonomy. WH Freeman, San Francisco
Sokal RR, Rohlf FJ (1995) Biometry: the principles and practice of statistics in biological research, 3rd edn. WH Freeman, New York
St Clair JB, Kleinschmit J (1986) Genotype-environment interaction and stability in ten-year height growth on Norway spruce clones (Picea abies Karst.). Silvae Genet 35:177–186
Wricke G (1962) Ueber eine Methode zur Enfassung der Oekologischen Streubreite in Feldversuchen. Z Pflanzenzuecht 47:92–96
Yamada Y (1962) Genotype by environment interaction and genetic correlation of the same trait under different environments. Jpn J Genet 37:498–509
Zobel BJ, Talbert J (1984) Applied forest tree improvement. Wiley, New York, p 505
Acknowledgements
The contributions from the following persons and institutions are gratefully acknowledged: Jochen Schmidt and Joseph Svolba helped at the initial stages of the clonal propagation and at the establishment and maintenance of the test sites; Dr. Tony (C.J.A.) Shelbourne, Dr. Steve McKeand and an anonymous reviewer provided constructive criticisms on the manuscript; the Alexander von Humboldt Foundation supported Kani Isik during the evaluation of the study; Dr. Wilfried Steiner provided a peaceful and friendly working environment at the LSFRI, Department of Forest Genetic Resources, Escherode, Germany, where the study has been performed.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D.B. Neale
Rights and permissions
About this article
Cite this article
Isik, K., Kleinschmit, J. Similarities and effectiveness of test environments in selecting and deploying desirable genotypes. Theor Appl Genet 110, 311–322 (2005). https://doi.org/10.1007/s00122-004-1840-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-004-1840-4