Abstract
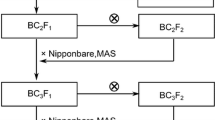
Morphogenetic processes in sequentially growing leaves and tiller buds are highly synchronized in rice (Oryza sativa L.). Consequently, the appearance of successive leaves in the main tiller acts as the ‘pacemaker’ for the whole shoot system development. The time interval between the appearance of successive leaves (days/leaf) in the main tiller is called the ‘phyllochron’. The objectives of the investigation reported here were: (1) to identify quantitative trait loci (QTLs) that control rice phyllochron and (2) to understand the roles of phyllochron QTLs as an underlying developmental factor for rice tillering. For this purpose we developed a set of recombinant inbred lines derived from a cross between IR36 (indica) and Genjah Wangkal (tropical japonica). Composite interval mapping detected three phyllochron QTLs located on chromosomes 4, 10 and 11, where the presence of a Genjah Wangkal allele increased phyllochron. The largest QTL (on chromosome 4) was located on the genomic region syntenic to the vicinity of the maize Teopod 2 mutation, while the QTL on chromosome 10 was close to the rice plastochron 1 mutation. These three phyllochron QTLs failed to coincide with major tiller number QTLs. However, one tiller number QTL was associated with small LOD peaks for phyllochron and tiller-bud dormancy that were linked in coupling phase, suggesting that linked small effects of phyllochron and tiller-bud dormancy might result in a multiplicative effect on tiller number.




Similar content being viewed by others
References
Ahn S, Tanksley SD (1993) Comparative linkage maps of the rice and maize genomes. Proc Natl Acad Sci USA 90:7980–7984
Ahn BO, Miyoshi K, Itoh J-I, Nagato Y, Kurata N (2002) A genetic and physical map of the region containing PLASTOCHRON1, a heterochronic gene, in rice (Oryza sativa L.). Theor Appl Genet 105:654–659
Araki E, Ebron LA, Cuevas RP, Mercado-Escueta D, Khush GS, Sheehy JE, Kato H, Fukuta Y (2003) Identification of low tiller gene in rice cultivar Aikawa1. Breed Res 5[Suppl 1]:95
Chen X, Temnykh S, Xu Y, Cho YG, McCouch SR (1997) Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L.). Theor Appl Genet 95:553–567
Dofing SM (1999) Inheritance of phyllochron in barley. Crop Sci 39:334–337
Friend DJC (1965) Tillering and leaf production in wheat as affected by temperature and light intensity. Can J Bot 43:1063–1076
Goto Y, Hoshikawa K (1989) Tillering behavior in Oryza sativa L. V. Analysis of varietal differences in tillering patterns (in Japanese). Jpn J Crop Sci 58:520–529
Gould SJ (1982) Change in development timing as a mechanism of macroevolution. In: Bonner JT (ed) Evolution and development. Springer, Berlin Heidelberg New York, pp 333–346
Haun JR (1973) Visual quantification of wheat development. Agron J 65:116–119
Hay RKM, Kirby, EJ (1991) Convergence and synchrony: a review of the coordination of development in wheat. Aust J Agric Res 42:661–700
Khush GS (2000) New plant type of rice for increasing the genetic yield potential. In: Nanda JS (ed) Rice breeding and genetics. Science Publishers, New Hampshire, pp 99–108
Klepper B, Rickman RW, Peterson CM (1982) Quantitative characterization of vegetative development in small cereal grains. Agron J 74:789–792
Kurata N, Nagamura Y, Yamamoto K, Harushima Y, Sue N, Wu J, Antonio BA, Shomura A, Shimizu T, Lin SY, Inoue T, Fukuda A, Shimano T, Kuboki Y, Toyama T, Miyamoto Y, Kirihara T, Hayasaka K, Miyao A, Monna L, Zhong HS, Tamura Y, Wang ZX, Momma T, Umehara Y, Yano M, Sasaki T, Minobe Y (1994) A 300-kilobase-interval genetic map of rice including 883 expressed sequences. Nat Genet 8:365–372
Lander ES, Green P, Abrahamson, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987)mapmaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Li JX, Yu SB, Xu CG, Tan YF, Gao YJ, Li XH, Zhang Q (2000) Analyzing quantitative trait loci for yield using a vegetatively replicated F2 population from a cross between the parents of an elite rice hybrid. Theor Appl Genet 101:248–254
Liao CY, Wu P, Hu B, Yi KK (2001) Effects of genetic background and environment on QTLs and epistasis for rice (Oryza sativa L.) panicle number. Theor Appl Genet 103:104–111
Luo LJ, Li ZK, Mei HW, Shu QY, Tabien R, Zhong DB, Ying CS, Stansel JW, Khush GS, Paterson AH (2001) Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. II. Grain yield components. Genetics 158:1755–1771
McCouch SR, Kochert G, Wang ZH, Khush GS, Coffman WR, Tanksley SD (1988) Molecular mapping of rice chromosomes. Theor Appl Genet 76:815–829
McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M, Fu B, Maghirang R, Li Z, Xing Y, Zhang Q, Kono I, Yano M, Fjellstrom R, DeClerck G, Schneider D, Cartinhour S, Ware D, Stein L (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9:199–207
McMaster GS, Klepper B, Rickman RW, Wilhelm WW, Willis WO (1991) Simulation of aboveground vegetative development and growth of unstressed winter wheat. Ecol Model 53:189–204
Mitchell KJ (1953) Influence of light and temperature on the growth of ryegrass (Lolium spp.). I. Pattern of vegetative development. Physiol Plant 6:21–46
Moore KJ, Moser LE (1995) Quantifying developmental morphology of perennial grasses. Crop Sci 35:37–43
Nemoto K, Morita S, Baba T (1995) Shoot and root development in rice related to the phyllochron. Crop Sci 35:24–29
Poethig RS (1988) Heterochronic mutations affecting shoot development in maize. Genetics 119:959–973
Ray JD, Yu L, McCouch SR, Champoux MC, Wang G, Nguyen HT (1996) Mapping quantitative trait loci associated with root penetration ability in rice (Oryza sativaL.). Theor Appl Genet 92:627–636
Veit B, Briggs SP, Schmidt RJ, Yanofsky MF, Hake S (1998) Regulation of leaf initiation by the terminal ear 1 gene of maize. Nature 393:166–168
Wang S, Basten CJ, Zeng Z-B (2001–2003) Windows qtl cartographer 2.0. Department of Statistics, North Carolina State University, Raleigh, N.C. (http://statgen.ncsu.edu/qtlcart/WQTLCart.htm)
Yan JQ, Zhu J, He CX, Benmoussa M, Wu P (1998) Quantitative trait loci analysis for the developmental behavior of tiller number in rice (Oryza sativa L.). Theor Appl Genet 97:267–274
Yin X, Kropff MJ (1996) The effect of temperature on leaf appearance in rice. Ann Bot 77:215–221
Zhu H, Qu F, Zhu LH (1993) Isolation of genomic DNAs from plants, fungi and bacteria using benzyl chloride. Nucleic Acids Res 21:5279–5280
Zhuang JY, Fan YY, Rao ZM, Wu JL, Xia YW, Zheng KL (2002) Analysis on additive effects and additive-by-additive epistatic effects of QTLs for yield traits in a recombinant inbred line population of rice. Theor Appl Genet 105:1137–1145
Acknowledgements
We thank Dr. Hideshi Yasui (Kyushu University) and Dr. Masahiro Yano (National Institute of Agrobiological Sciences) for valuable suggestions on experimental design; Prof. Hirohisa Kishino (The University of Tokyo) for his kind guidance for multiple-regression analysis; Dr. Ma. Rebecca C. Laza (The International Rice Research Institute) for critically reviewing the manuscript; Dr. M.T. Jackson (IRRI) for kindly providing the seeds of Genjah Wangkal rice; Mrs. Mitsuko Konno (The University of Tokyo) for her technical support. This work was supported by Grant-in-Aid for Scientific Research (nos.12660012 and15380013 to K.N.) from the Ministry of Education, Science, Sports and Culture, Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D.J. Mackill
Rights and permissions
About this article
Cite this article
Miyamoto, N., Goto, Y., Matsui, M. et al. Quantitative trait loci for phyllochron and tillering in rice. Theor Appl Genet 109, 700–706 (2004). https://doi.org/10.1007/s00122-004-1690-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-004-1690-0