Abstract
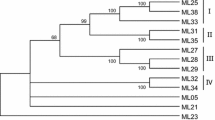
Degenerate primers based on conserved regions of the nucleotide binding site (NBS) domain (encoded by the largest group of cloned plant disease resistance genes) were used to isolate a set of 15 resistance gene analogs (RGA) from the diploid species Avena strigosa Schreb. These were grouped into seven classes on the basis of 60% or greater nucleic acid sequence identity. Representative clones were used for genetic mapping in diploid and hexaploid oats. Two RGAs were mapped at two loci of the linkage group AswBF belonging to the A. strigosa × A. wiestii Steud map, and ten RGAs were mapped at 15 loci in eight linkage groups belonging to the A. byzantina C. Koch cv. Kanota × A. sativa L. cv. Ogle map. A similar approach was used for targeting genes encoding receptor-like kinases. Three different sequences were obtained and mapped to two linkage groups of the hexaploid oat map. Associations were explored between already known disease resistance loci mapped in different populations and the RGAs. Molecular markers previously linked to crown rust and barley yellow dwarf resistance genes or quantitative trait loci were found in the Kanota × Ogle map linked to RGAs at a distance ranging from 0 cM to 20 cM. Homoeologous RGAs were found to be linked to loci either conferring resistance to different isolates of the same pathogen or to different pathogens. This suggests that these RGAs identify genome regions containing resistance gene clusters.



Similar content being viewed by others
References
Aarts MGM, Lintel Hekkert B, Holub EB, Beynon JK, Stiekema WJ, Pereira A (1998) Identification of R-gene homologous DNA fragments genetically linked to disease resistance loci in Arabidopsis thaliana. Mol Plant Microbe Interact 11:251–258
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res 25:2389–3402
Backer B, Zambryski P, Staskawicz B, Dinesh-Kumar SP (1997) Signaling in plant-microbe interactions. Science 276:726–733
Backes G, Madsen LH, Jaiser H, Stougaard J, Herz M, Mohler V, Jahoor A (2003) Localisation of genes for resistance against Blumeria graminis f. sp. hordei and Puccinia graminis in a cross between a barley cultivar and a wild barley (Hordeum vulgare ssp. Spontaneum) line. Theor Appl Genet 106:353–362
Bai J, Pennill LA, Ning J, Lee SW, Ramalingam J, Webb CA, Zhao B, Sun Q, Nelson JC, Leach JE, Hulbert SH (2002) Diversity in nucleotide binding site-leucine rich repeat genes in cereals. Genome Res 12:1871–1884
Barbosa-Neto JF, Siripoonwiwat W, O’Donoughue LS, Gray SM, Smith DM, Kolb FL, Gourmet C, Brown CM, Sorrells ME (2000) Chromosomal regions associated with yellow dwarf virus resistance in oat. Euphytica 114:67–76
Baudino S, Hansen S, Brettschneider R, Hecht VFG, Dresselhaus T, Lörz H, Dumas C, Rogowsky PM (2001) Molecular characterisation of two novel maize LRR receptor-like kinases, which belong to the SERK gene family. Planta 213:1–10
Burnet PA (1983) Preface. Barley Yellow Dwarf Workshop, Mexico, pp 6–13
Bush AL, Wise RP (1996) Crown rust resistance loci on linkage groups 4 and 13 in cultivated oat. J Hered 87:427–432
Bush AL, Wise RP (1998) High-resolution mapping adjacent to the Pc71 crown-rust resistance locus in hexaploid oat. Mol Breed 4:13–21
Bush AL, Wise RP, Rayapati PJ, Lee M (1994) Restriction fragment length polymorphisms linked to genes for resistance to crown rust (Puccinia coronata) in near-isogenic lines of hexaploid oat (Avena sativa). Genome 37:823–831
Cannon SB, Zhu H, Baumgarten AM, Spangler R, May G, Cook DR, Young ND (2002) Diversity, distribution, and ancient taxonomic relationships within the TIR and Non-TIR NBS-LRR resistance gene subfamilies. J Mol Evol 54:548–562
Chen G, Portyanko VA, Rines HW, Phillips RL, Leonard KJ, Ochocki GE, Stuthman DD (2000) Identification of QTLs for partial resistance to crown rust of oats (abstract). In: ASA-CSSA-SSA (ed) ASA-CSSA-SSA Annu Meet. ASA-CSSA-SSA, Madison, Wis., p 187
Cheng DW, Armstrong KC, Tinker N, Wight CP, He S, Lybaert A, Fedak G, Molnar SJ (2002) Genetic and physical mapping of Lrk10-like receptor kinase sequences in hexaploid oat (Avena sativa L.). Genome 45:100–109
Chong J, Howes NK, Brown PD, Harder DE (1994) Identification of the stem rust resistance gene Pg9 and its association with crown rust resistance and endosperm proteins in Dumont oat. Genome 37:440–447
Collins NC, Webb CA, Seah S, Ellis JG, Hulbert SH, Pryor A (1998) The isolation and mapping of disease resistance gene analogs in maize. Mol Plant Microbe Interact 11:968–978
Deng Z, Gmitter FG Jr (2003) Cloning and characterization of receptor kinase class disease resistance gene candidates in Citrus. Theor Appl Genet 108:53–61
Donald TM, Pellerone F, Adam-Blondon AF, Bouquer A, Thomas MR, Dry IB (2002) Identification of resistance gene analogs linked to a powdery mildew resistance locus in grapevine. Theor Appl Genet 104:610–618
Feuillet C, Schachermayr G, Keller B (1977) Molecular cloning of a new receptor-like kinase gene encoded at the Lr10 disease resistance locus of wheat. Plant J 11:45–52
Forsberg RA (1990) The use of monosomic alien substitution lines in interploidy gene transfer in Avena. Bulg J Biotechnol 4:27–30
Fourmann M, Charlot F, Froger N, Delourne R, Brunel D (2001) Expression, mapping, and genetic variability of Brassica napus disease resistance gene analogues. Genome 44:1083–1099
Halterman D, Zhou F, Wei F, Wise RP, Schulze-Lefert P (2001) The MLA6 coiled-coil, NBS-LRR protein confers AvrMla6-dependent resistance specificity to Blumeria graminis f. sp. hordem in barley and wheat. Plant J 25:335–348
Hammond-Kosack KE, Jones JJ (1997) Plant disease resistance genes. Annu Rev Plant Physiol Plant Mol Biol 48:575–607
Harder DE (1994) Virulence dynamics of Puccinoa graminis f. sp. Avenae in Canada, 1921–1993. Phytopathology 84:739–746
Harder DE, Haber S (1992) Oat disease and pathologic techniques. Oat Sci Technol Agron Monogr No 33:307–425
Higgins DG, Sharp PM (1988) clustal w: a package for performing multiple sequence alignment on a microcomputer. Gene 73:237–244
Holland JB, Munkvold GP (2001) Genetic relationships of crown resistance, grain yield, test weight, and seed weight in oat. Crop Sci 41:1041–1050
Huettel B, Santra D, Muelbauer FJ, Kahl G (2002) Resistance gene analogues of chickpea (Cicer arietinum L.): isolation, genetic mapping and association with a Fusarium resistance gene cluster. Theor Appl Genet 105:479–490
Hulbert SH, Webb CA, Smith SM, Sun Q (2001) Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol 39:285–312
Jin H, Domier LL, Kolb FL, Brown CM (1998) Identification of quantitative loci for tolerance to barley dwarf virus in oat. Phytopathology 88:410–415
Kanazin V, Marek LF, Shoemaker RC (1996) Resistance gene analogs are conserved and clustered in soybean. Proc Natl Acad Sci USA 93:11746–11750
Kremer CA, Lee M, Holland JB (2001) A restriction fragment length polymorphism based linkage map of a diploid Avena recombinant inbred line population. Genome 44:192–204
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) mapmaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Leister D, Ballvora A, Salamini F, Gebhardt C (1996) A PCR-based approach for isolating pathogen resistance genes from potato with potential for wide application in plants. Nat Genet 14:421–429
Madsen LH, Collins NC, Rakwalska M, Backes G, Sandal N, Krusell L, Jensen J, Waterman EH, Jahoor A, Ayliffe M, Pryor A, Langridge P, Schulze-Lefert P, Stougaard J (2003) Barley disease resistance gene analogs of the NBS-LRR class: identification and mapping. Mol Genet Genomics 269:150–161
Marshall HG, Shaner GE (1992) Genetics and inheritance in oat. In: Marshall HG, Sorrells ME (eds) Oat science and technology. American Society of Agronomy and Crop Science Society of America, Madison, pp 510–571
Melotto M, Kelly JD (2001) Fine mapping of the Co-4 locus of common bean reveals a resistance gene candidate, COK-4, that encodes for a protein kinase. Theor Appl Genet 103:508–517
Meyers BC, Chin DB, Shen KA, Sivaramakrishanan S, Lavelle DO, Zhang ZM Michelmore RW (1998) The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell 10:1817–1832
Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20:317–332
Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15:809–834
Mohler V, Klarhr A, Wenzel G, Schwarz G (2002) A resistance gene analog useful for targeting disease resistance genes against different pathogens on group 1S chromosomes of barley, wheat and rye. Theor Appl Genet 105:364–368
Myers EW, Miller W (1988) aling: optimal alignments in linear space. Comp Appl Biosci 4:11–17
Noir S, Combes MC Anthony F, Lashermes P (2001) Origin, diversity and ecolution of NBS-type disease-resistance gene homologues in coffee trees (Coffea l.). Mol Genet Genomics 265:654–662
O’Donoughue LS, Kianian SF, Rayapati PJ, Penner GA, Sorrells ME, Tanksley SD, Phillips RL, Rines HW, Lee M, Fedak G, Molnar SJ, Hoffman D, Salas CA, Wu B, Autrique E, van Deynze A (1995) A molecular linkage map of cultivated oat. Genome 38:368–380
O’Donoughue LS, Chong J, Wight CP, Fedak G, Molnar SJ (1996) Localization of stem rust resistance genes and associated molecular markers in cultivates oat. Phytopathology 86:719–727
Pan Q, Wendel J, Fluhr R (2000) Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. J Mol Evol 50:203–213
Peñuela S, Danesh D, Young ND (2002) Targeted isolation, sequence analysis, and physical mapping of nonTIR NBS-LRR genes in soybeans. Theor Appl Genet 104:261–272
Portyanko VA, Hoffman DL, Lee M, Holland JB (2001) A linkage map of hexaploid oat based on grass anchor DNA clones and its relationship to other oat maps. Genome 44:249–265
Radwan O, Bouzidi MF, Vear F, Philippon J, Tourvieille de Labrouhe D, Nicolas P, Mouzeyar S (2003) Identification of non-TIR-NBS-LRR markers linked to the PI5/PI8 locus for resistance to downy mildew in sunflower. Theor Appl Genet 106:1438–1446
Rajhathy T, Thomas H (1974) Cytogenetics of oats (Avena L.). Misc Publ Genet Soc Can 2:1–90
Rayapaty PJ, Gregory JW, Lee M, Wise RP (1994) A linkage map of diploid Avena based on RFLP loci and a locus conferring resistance to nine isolates of Puccinia coronata var. avenae. Theor Appl Genet 89:831–837
Robertson DS (1985) A possible technique for isolating genic DNA for quantitative traits in plants. J Theor Biol 117:1–10
Rooney WL, Rines HW, Phillips RL (1994) Identification of RFLP markers linked to crown rust resistance genes Pc91 and Pc92 in oat. Crop Sci 34:940–944
Rossi M, Araujo PG, Paulet F, Garsmeur P, Dias VM, Chen H, Van Sluys MA, D’Hont A (2003) Genomic distribution and characterization of EST-derived resistance gene analogs (RGAs) in sugarcane. Mol Genet Genomics. DOI 10.1007/s00438-003-0849-8
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Salmeron J, Oldroyd GED, Rommens CM, Scofield SR, Kim HS, Lavelle DT, Dahlbeck D, Staskawicz BJ (1996) Tomato Prf is a member of leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase cluster. Cell 86:123–133
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Seah S, Bariana H, Jahier J, Sivasithamparam K, Lagudah ES (2001) The introgressed segment carrying rust resistance genes Yr17, Lr37 and Sr38 wheat can be assayed by a cloned disease resistance gene-like sequence. Theor Appl Genet 102:600–605
Sebesta J, Roderick HW, Stojanovic S, Zwatz B, Harder DE, Corazza L (2000) Genetic basis of oat resistance to fungal diseases. Plant Protect Sci 36:23–38
Sharp PJ, Kreiss M, Shewry P, Gale MD (1988) Location of β-amylase sequences in wheat and its relatives. Theor Appl Genet 75:289–290
Shen KA, Meyers BC, Islam Faridi NM, Chin DB, Stelly DM, Michelmore RW (1998) Resistance gene candidates identified by PCR with degenerate oligonucleotide primers map to clusters of resistance genes in lettuce. Mol Plant Microbe Interact 11:815–823
Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Gardner J, Wang B, Holsten TE, Zhai WX, Zhu LH, Fauquet X, Ronald PC (1995) A receptor kinase-like protein encoded by the rice disease resistance gene Xa21. Science 270:1804–1806
Tinker N (2003) Oats mapping database. http://www.gnomad.agr.ca
Van der Vossen EA, van der Voort JN, Kanyuka K, Bendahmane A, Snedbrink H, Baulcombe DC, Bakker J, Stiekema WJ, Klein-Lankhorst RM (2000) Homologues of a single resistance-gene cluster in potato confer resistance to distinct pathogens: a virus and a nematode. Plant J 23:567–576
Wei F, Gobelman-Werner K, Morrol SM, Kurth J, Mao L, Wing R, Leister D, Schulze-Lefert P, Wise RP (1999) The Mla (powdery mildew) resistance cluster is associated with three NBS-LRR gene families and suppressed recombination within a 240-kb DNA interval on chromosome 5S (1HS) of barley. Genetics 153:1929–1948
Wight CP, Tinker NA, Kianian SF, Sorrells ME, O’Donoghue LS, Hoffman DL, Groh S, Scoles GJ, Li CD, Webster FH, Phillips RL, Rines HW, Livingston SM, Armstrong KC, Fedak G, Molnar SJ (2003) A molecular marker map in ‘Kanota’ × ‘Ogle’ hexaploid oat (Avena spp) enhanced by additional markers and a robust framework. Genome 46:28–47
Wight CP, O’Donoughue LS, Chong J, Tinker NA, Molnar SJ (2004) Discovery, localization, and sequence characterization of molecular markers for the crown rust genes Pc38, Pc39, and Pc48 in cultivated oat (Avena sativa L.). Mol Breed (in press)
Yu GX, Wise RP (2001) An anchored AFLP- and retrotransposon-based map of diploid Avena. Genome 43:736–749
Yu YG, Buss GR, Saghai-Maroof MA (1996) Isolation of a superfamily of candidate disease-resistance genes in soybean Based on a conserved nucleotide-binding site. Proc Natl Acad Sci USA 93:11751–11756
Zhang X (1998) Leucine-rich repeat receptor-like kinases in plants. Plant Mol Biol Report 16:301–311
Zhu S, Kaeppler HF (2003) Identification of quantitative trait loci for resistance to crown rust in oat line MAM17–5. Crop Sci 43:358–366
Zhu S, Kolb FL, Kaeppler HF (2003) Molecular Mapping of genomic regions underlying barley yellow dwarf tolerance in cultivated oat (Avena sativa L.) Theor Appl Genet 106:1300–1306
Acknowledgements
We thank Dr. Mike Lee (Iowa State University, USA) for his kind contribution in mapping the diploid population. We also thank Drs. Ken Armstrong, Nick Tinker and Charlene Wight (Agriculture and Agri-Food, Canada) for providing us with the Kanota × Ogle seeds, for sharing unpublished data, and for helpful advice. Dr. Howard Rines (University of Minnesota, USA) is thanked for sharing preliminary data, and Eva Friero for technical assistance. This work was financed by the Spanish Ministry of Science and Technology (AG1999-0918).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Langridge
Rights and permissions
About this article
Cite this article
Irigoyen, M.L., Loarce, Y., Fominaya, A. et al. Isolation and mapping of resistance gene analogs from the Avena strigosa genome. Theor Appl Genet 109, 713–724 (2004). https://doi.org/10.1007/s00122-004-1679-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-004-1679-8