Abstract
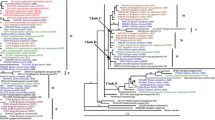
Species of the genus Elymus are closely related to some important cereal crops and may thus serve as potential alien genetic resources for the improvement of these crops. E. humidus is indigenous to Japan and is well adapted to a humid climate. However, the phylogenetic and evolutionary relationships between E. humidus and other Elymus species are unclear. To elucidate these relationships, we examined the sequences of three non-coding regions of chloroplast DNA (cpDNA) and the amplified fragment length polymorphism (AFLP) variation of nuclear DNA in E. humidus and other related species. A total of 15 sequence mutations from the three non-coding regions, trnL-trnF, trnF-ndhJ(C), and atpB-rbcL, covering approximately 1,800 bp, were detected in the Elymus species. A phylogenic tree resulting from the cpDNA sequence data revealed that all the species containing the St nuclear genome (St, StH, StY, and StHY) formed a well-supported clade that is remote from the Hordeum species (H). This result strongly supports the finding that Pseudoroegneria is the maternal genome donor to the genus Elymus. In addition, E. humidus showed the closest relationship with the cpDNA genome of the Pseudoroegneria species. The AFLP analysis detected 281 polymorphic bands with 11 AFLP primer combinations. The AFLP result showed that E. humidus is relatively closer to E. tsukushiensis. However, the cpDNA sequencing results indicated that E. humidus and E. tsukushiensis have different cytoplasmic origins. Our results suggest that the evolutionary process between E. humidus and E. tsukushiensis is not monophyletic, although the two species have similar morphological characters and adaptability.


Similar content being viewed by others
References
Ban T (1997) Evaluation of resistance of Fusarium head blight in indigenous Japanese species of Agropyron (Elymus). Euphytica 97:39–44
Cipriani G, Testolin R, Gardner R (1998) Restriction-site variation of PCR-amplified chloroplast DNA regions and its implication for the evolution of Actinidia. Theor Appl Genet 96:389–396
Dewey DR (1984) The genomic system of classification as a guide to intergeneric hybridization with the perennial Triticeae. In: Gustafson JP (ed) Gene manipulation in plant improvement. Plenum, New York, pp 209–280
Doyle JJ, Doyle JL (1990) Isolate of plant DNA from fresh tissue. Focus 12:13–15
Felsenstein, J (1995) PHYLIP: phylogeny inference package, ver 3.57c. University of Washington Press, Seattle
Jensen KB (1990) Cytology and taxonomy of E. grandiglumis, E. alatavicus, and E. batalinii (Poaceae: Triticeae). Genome 33:668–673
Le Thierry d’Ennequin M, Panaud O, Toupance B, Sarr A (2000) Assessment of genetic relationships between Setaria italica and its wild relative S. viridis using AFLP markers. Theor Appl Genet 100:1061–1066
Mackill DJ, Zhang Z, Redoña ED, Colowit PM (1996) Level of polymorphism and genetic mapping of AFLP markers in rice. Genome 39:969–977
Manen JF, Natall A (1995) Comparison of the evolution of ribulosee-1,5-bisphosphate carboxylase (rbcL) and atpB-rbcL non-coding spacer sequences in a recent plant group, the tribe Rubieae (Rubiaceae). J Mol Evol 41:920–927
Mason-Gamer RJ, Kellogg EA (1996) Testing for phylogenetic conflict among molecular data sets in the tribe Triticeae (Gramineae). Syst Biol 45:524–545
Mason-Gamer RJ, Orme NL, Anderson CM (2002) Phylogenetic analysis of North American Elymus and the monogenomic Triticeae (Poaceae) using three chloroplast DNA data sets. Genome 45:991–1002
McDade LA, Moody ML (1999) Phylogenetic relationship among Acanthaceae: evidence from noncoding trnL-trnF chloroplast DNA sequences. Am J Bot 86:70–80
McMillan E, Sun G. (2003) Genetic relationship of tetraploid Elymus species and their donor species inferred from polymerase chain reaction-restriction length polymorphism analysis of chloroplast gene regions. Theor Appl Genet (in press). DOI 10.1007/s00122-003-1453-3
Ogihara Y, Tsunewaki K (1988) Diversity and evolution of chloroplast DNA in Triticum and Aegilops as revealed by restriction fragment analysis. Theor Appl Genet 76:321–332
Ogihara Y, Isono K, Kojima T, Endo A, Hanaoka M, Shiina T, Terachi T, Utsugi S, Murata M, Mori N, Takumi S, Ikeo K, Gojobori T, Murai R, Murai K, Matsuoka Y, Ohnishi Y, Tajiri H, Tsunewaki K (2000) Chinese spring wheat (Triticum aestivum L.) chloroplast genome: complete sequence and contig clones. Plant Mol Biol Rep 18:243–253
Olmstead RG, Palmer JD (1994) Chloroplast DNA systematic: a review of method and data analysis. Am J Bot 81:1205–1224
Redingbaugh MG, Jones TA, Zhang Y (2000) Ubiquity of the St chloroplast genome in St-containing Triticeae polyploids. Genome 43:846–852
Sakamoto S (1966) Cytogenetic studies in the tribe Triticeae. IV. Natural hybridization among Japanese Agropyron species. Jpn J Genet 41:189–201
Sakamoto S, Matsumura M (1966) Cytogenetic studies in the tribe. II. Tetraploid and hexaploid hybrid of Agropyron. Jpn J Genet 41:155–168
Sasanuma T, Endo TR, Ban T (2002) Genetic diversity of three Elymus species indigenous to Japan and East Asia (E. tsukushiensis, E. humidus, and E. dahuricus) detected by AFLP. Genes Genet Syst 77:429–438
Scotti I, Mariani A, Verona V, Candolini A, Cenci CA, Olivieri AM (2002) AFLP markers and cytoaxonomic analysis reveal hybridization in the genus Schoenus (Cyperaceae). Genome 45:222–228
Stebbins GL, Valencia JI, Valencia RM (1946) Artificial and natural hybrids in the Gramineae, tribe Hordeae. II. Agropyron, Elymus, and Hordeum. Am J Bot 33:579–586
Tsunewaki K, Ogihara Y (1983) The molecular basis of genetic diversity among cytoplasms of Triticum and Aegilops species. II. On the origin of polyploid wheat cytoplasms as suggested by chloroplast DNA restriction fragment patterns. Genetics 104:155–171
Wang XE, Chen PD, Liu DJ, Zhang P, Zhou B, Friebe B, Gill BS (2001) Molecular cytogenetic characterization of Roegneria ciliaris chromosome additions in common wheat. Theor Appl Genet 102:651–657
Wolfe AD, Elisens WJ, Watson LE, Depamphilis CW (1997) Using restriction-site variation of PCR-amplified cpDNA genes for phylogenetic analysis of Tribe Cheloneae (Scrophulariaceae). Am J Bot 84:555–564
Xu DH, Abe J, Sakai M, Kanazawa A, Shimamoto Y (2000) Sequence variation of non-coding regions of chloroplast DNA of soybean and related wild species and its implications for the evolution of different chloroplast haplotypes. Theor Appl Genet 101:724–732
Xu DH, Abe J, Kanazawa A, Gai JY, Shimamoto Y (2001) Identification of sequence variations by PCR-RFLP and its application to the evaluation of cpDNA diversity in wild and cultivated soybeans. Theor Appl Genet 102:683–688
Acknowledgements
We thank Dr. Richard R.C. Wang (USDA-ARS, Forage and Range Research Laboratory, Utah State University, USA), Professor R. von Bothmer (Department of Plant Breeding, The Swedish University of Agricultural Sciences, Sweden), and Dr. N. Watanabe for (Faculty of Agriculture, Gifu University, Japan) for providing plant materials. Thanks are also due to Dr. H. Tsunematsu (Japan International Research Center for Agriculture Sciences, Japan) for his many useful comments on this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Dvorak
Rights and permissions
About this article
Cite this article
Xu, D.H., Ban, T. Phylogenetic and evolutionary relationships between Elymus humidus and other Elymus species based on sequencing of non-coding regions of cpDNA and AFLP of nuclear DNA. Theor Appl Genet 108, 1443–1448 (2004). https://doi.org/10.1007/s00122-004-1588-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-004-1588-x