Abstract
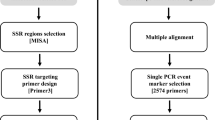
A set of 398 simple sequence repeat markers (SSRs) have been developed and characterised for use with genetic studies of Brassica species. Small-insert (250–900 bp) genomic libraries from Brassica rapa, B. nigra, B. oleracea and B. napus, highly enriched for dinucleotide and trinucleotide SSR motifs, were constructed. Screening the clones with a mixture of oligonucleotide repeat probes revealed positive hybridisation to between 75% and 90% of the clones. Of these, 1,230 were sequenced. Primer pairs were designed for 398 SSR clones, and of these, 270 (67.8%) amplified a PCR product of the expected size in their focal and/or closely related species. A further screen of 138 primers pairs that produced a PCR product in B. napus germplasm found that 86 (62.3%) revealed length polymorphisms within at least one line of a test array representing the four Brassica species. The results of this screen were used to identify 56 SSRs and were combined with 41 SSRs that had previously shown polymorphism between the parents of a B. napus mapping population. These 97 SSR markers were mapped relative to a framework of RFLP markers and detected 136 loci over all 19 linkage groups of the oilseed rape genome.



Similar content being viewed by others
References
Axelsson T, Bowman CM, Sharpe AG, Lydiate DJ, Lagercrantz U (2000) Amphidiploid Brassica juncea contains conserved progenitor genomes. Genome 43:679–688
Bohuon EJR, Keith DJ, Parkin IAP, Sharpe AG, Lydiate DJ (1996) Alignment of the conserved C genomes of Brassica oleracea and Brassica napus. Theor Appl Genet 93:833–839
Cavell AC, Lydiate DJ, Parkin IAP, Dean C, Trick M (1998) Collinearity between a 30 centimorgan segment of Arabidopsis thaliana chromosome 4 and duplicated regions within the Brassica napus genome. Genome 41:62–69
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Edwards KJ, Barker JHA, Daly A, Jones C, Karp A (1996) Microsatellite libraries enriched for several microsatellite sequences in plants. BioTechniques 20:759–760
Goldstein DB, Schlötterer C (1999) Microsatellites: evolution and applications. Oxford University Press, Oxford
Howell EC, Barker GC, Jones GH, Kearsey MJ, King GJ, Kop EP, Ryder CD, Teakle GR, Vicente JG, Armstrong SJ (2002) Integration of the cytogenetic and genetic linkage maps of Brassica oleracea. Genetics 161:1225–1234
Khlestkina EK, Pestsova EG, Salina E, Roder MS, Arbuzova VS, Koval SF, Borner A (2002) Genetic mapping and tagging of wheat genes using RAPD, STS and SSR markers. Cell Mol Biol Lett 7:795–802
Kowalski SP, Lan T-H, Feldmann KA, Paterson AH (1994) Comparative mapping of Arabidopsis thaliana and Brassica oleracea chromosomes reveals islands of conserved organization. Genetics 138:1–12
Lagercrantz U, Lydiate DJ (1995) RFLP mapping in Brassica nigra indicates differing recombination rates in male and female meioses. Genome 38:255–264
Lagercrantz U, Lydiate DJ (1996) Comparative genome mapping in Brassica. Genetics 144:1903–1910
Lincoln SE, Daly MJ, Lander ES (1987) MAPMAKER v3.0. A computer programme. Whitehead Institute, Cambridge, Mass.
Lionneton E, Ravera S, Sanchez L, Aubert G, Delourme R, Ochatt S (2002) Development of an AFLP-based linkage map and localization of QTLs for seed fatty acid content in condiment mustard (Brassica juncea) Genome 45:1203–1215
Lombard V, Delourme R (2001) A consensus linkage map for rapeseed (Brassica napus L.): construction and integration of three individual maps from DH populations. Theor Appl Genet 103:491–507
Lowe AJ, Jones AE, Raybould AF, Trick M, Moule CJ, Edwards KJ (2002) Transferability and genome specificity of a new set of microsatellite primers among Brassica species of the U triangle. Mol Ecol Notes 2:7–11
Lydiate D, Sharpe A, Largercrantz U, Parkin I (1993) Mapping the Brassica genome. Outl Agric 22:85–92
Parkin IAP, Sharpe AG, Keith DJ, Lydiate DJ (1995) Identification of the A and C genomes of amphidiploid Brassica napus (oilseed rape). Genome 38:1122–1131
Plieske J, Struss D (2001) Microsatellite markers for genome analysis in Brassica. I. Development in Brassica napus and abundance in Brassicaceae species. Theor Appl Genet 102:689–694
Rae SJ, Aldam C, Dominguez I, Hoebrechts M, Barnes SR, Edwards KJ (2002) Development and incorporation of microsatellite markers into the linkage map of sugar beet (Beta vulgaris spp.) Theor Appl Genet 100:1240–1248
Saal B, Plieske J, Hu J, Quiros CF, Struss D (2001) Microsatellite markers for genome analysis in Brassica. II. Assignment of rapeseed microsatellites to the A and C genomes and genetic mapping in Brassica oleracea L. Theor Appl Genet 102:695–699
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Sebastian RL, Howell EC, King GJ, Marshall DF, Kearsey MJ (2000) An integrated AFLP and RFLP Brassica oleracea linkage map from two morphologically distinct doubled-haploid mapping populations. Theor Appl Genet 100:75–81
Sharopova N, McMullen MD, Schultz L, Schroeder S, Sanchez-Villeda H, Gardiner J, Bergstrom D, Houchins K, Melia-Hancock S, Musket T, Duru N, Polacco M, Edwards K, Ruff T, Register JC, Brouwer C, Thompson R, Velasco R, Chin E, Lee M, Woodman-Clikeman W, Long MJ, Liscum E, Cone K, Davis G, Coe EH (2002) Development and mapping of SSR markers for maize. Plant Mol Biol 48: 463–481
Sharpe AG, Parkin IAP, Keith DJ, Lydiate DJ (1995) Frequent non-reciprocal translocations in the amphidiploid genome of oilseed rape (Brassica napus). Genome 38:1112–1121
Soltis DE, Soltis PS (1995) The dynamic nature of polyploid genomes. Proc Natl Acad Sci USA 92:8089–8091
Song K, Lu P, Tang K, Osborn TC (1995) Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc Natl Acad Sci USA 92:7719–7723
Squirrell J, Hollingsworth PM, Woodhead M, Russell J, Lowe AJ, Gibby M, Powell W (2003) How much effort is required to isolate nuclear microsatellites from plants? Mol Ecol 12:1339–1348
Suwabe K, Iketani H, Nunome T, Kage T, Hirai M (2002) Isolation and characterization of microsatellites in Brassica rapa L. Theor Appl Genet 104:1092–1098
SzewcMcFadden AKS, Kresovich S, Bliek SM, Mitchell SE, McFerson JR (1996) Identification of polymorphic, conserved simple sequence repeats (SSRs) in cultivated Brassica species. Theor Appl Genet 93:534–538
Tang S, Yu JK, Slabaugh MB, Shintani DK, Knapp SJ (2002) Simple sequence repeat map of the sunflower genome. Theor Appl Genet 105:1124–1136
U N (1935) Genomic analysis of Brassica with special reference to the experimental formation of B. napus and its peculiar mode of fertilization. Jpn J Bot 7:389–452
Warwick SI, Black LD, Aguinagalde I (1992) Molecular systematics of Brassica and allied genera (Subtribe Brassicinae, Brassiceae) - chloroplast DNA variation in the genus Diplotaxis. Theor Appl Genet 83:839–850
Westman AL, Kresovich S (1998) The potential for cross-taxa simple-sequence repeat (SSR) amplification between Arabidopsis thaliana L. and crop brassicas. Theor Appl Genet 96:272–281
Westman AL, Kresovich S (1999) Simple sequence repeat (SSR)-based marker variation in Brassica nigra genebank accessions and weed populations. Euphytica 109:85–92
Zane L, Bargelloni L, Patarnello T (2002) Statregies for microsatellite isolation: a review. Mol Ecol 11:1–16
Acknowledgements
We would like to thank staff at IACR Long Ashton and the John Innes Centre who provided support and help during laboratory work. This work was primarily funded by a grant (ref D08078) from the Biotechnology and Biological Sciences Research Council. Supporting finance was provided by IACR Long Ashton, John Innes Centre, BBSRC and the Natural Environment Research Council.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by O. Savolainen
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Lowe, A.J., Moule, C., Trick, M. et al. Efficient large-scale development of microsatellites for marker and mapping applications in Brassica crop species. Theor Appl Genet 108, 1103–1112 (2004). https://doi.org/10.1007/s00122-003-1522-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-003-1522-7