Abstract
The risk of severe COVID-19 increases with age as older patients are at highest risk. Thus, there is an urgent need to identify how severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) interacts with blood components during aging. We investigated the whole blood transcriptome from the Genotype-Tissue Expression (GTEx) database to explore differentially expressed genes (DEGs) translated into proteins interacting with viral proteins during aging. From 22 DEGs in aged blood, FASLG, CTSW, CTSE, VCAM1, and BAG3 were associated with immune response, inflammation, cell component and adhesion, and platelet activation/aggregation. Males and females older than 50 years old overexpress FASLG, possibly inducing a hyperinflammatory cascade. The expression of cathepsins (CTSW and CTSE) and the anti-apoptotic co-chaperone molecule BAG3 also increased throughout aging in both genders. By exploring single-cell RNA-sequencing data from peripheral blood of SARS-CoV-2-infected patients, we found FASLG and CTSW expressed in natural killer cells and CD8 + T lymphocytes, whereas BAG3 was expressed mainly in CD4 + T cells, naive T cells, and CD14 + monocytes. In addition, T cell exhaustion was associated with increased expression of CCL4L2 and DUSP4 over blood aging. LAG3, PDCD1, TIGIT, VCAM1, HLA-DRA, and TOX also increased in individuals aged 60–69 years old; conversely, the RGS2 gene decreased with aging. We further identified a distinct gene expression profile associated with type I interferon signaling following blood aging. These results revealed changes in blood molecules potentially related to SARS-CoV-2 infection throughout aging, emphasizing them as therapeutic candidates for aggressive clinical manifestation of COVID-19.
Key messages
• Prediction of host-viral interactions in the whole blood transcriptome during aging.
• Expression levels of FASLG, CTSW, CTSE, VCAM1, and BAG3 increase in aged blood.
• Blood interactome reveals targets involved with immune response, inflammation, and blood clots.
• SARS-CoV-2-infected patients with high viral load showed FASLG overexpression.
• Gene expression profile associated with T cell exhaustion and type I interferon signaling were affected with blood aging.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Coronavirus disease 2019 (COVID-19) is a pandemic infection caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1]. This virus is associated with a broad spectrum of respiratory disturbances, varying from upper airway symptoms to aggressive pneumonia [2]. In lung parenchyma, it produces alveolar edema, fibrin deposition, and hemorrhage [2]. Notably, vascular changes are one of the distinctive features of COVID-19. Many patients have demonstrated clinical signs of thrombotic microangiopathy [3], with intravascular coagulation and thrombosis associated with multisystem organ failure [4].
Although SARS-CoV-2 affects the lungs primordially, there is convincing evidence that it alters the coagulation processes in severe cases [5]. The formation of blood clots disrupts circulation due to thrombosis, pulmonary embolism, and heart attacks [3, 4]. The association of these changes in coagulation and protein aggregates with alterations in inflammatory parameters results in increased COVID-19 mortality at alarming proportions. The micro-thrombotic environment arises from the hyperactivation of the coagulation cascade associated with hyperinflammation and immune activities [6]. Of note, the clotting process is complex and likely orchestrated by the massive release of pro-inflammatory mediators, cytokines, and tumor necrosis factor (TNF), mainly released from monocytes and endothelial cells [6]. The fact that the mortality rate of patients aged 60 years and over is higher than those under 60 years is indisputable [7]. Most of these critical cases are associated with the “cytokine storm,” as these exacerbated immune reactions may lead to the early death of elderly people regardless of comorbidities related to more severe cases [8].
Recent findings are correlating the blood subtype with viral susceptibility and infection [9, 10]. However, the individual molecular machinery of blood cells may be detrimental for the grade and type of response, such as exacerbated or reduced inflammation. Considering that aging is one of the most significant risk factors for severe cases of COVID-19, it is essential to determine blood host genetic variation throughout the aging process. The evolutionary conservation between the 2019 novel SARS-CoV-2 and SARS-CoV [11] allows us to understand similarities and differences between these coronaviruses into public databases. Furthermore, using computational predictions of SARS-CoV–human protein–protein interactions (PPIs), we can identify possible mechanisms behind the viral infection and potential drug targets [12, 13].
Considering that older individuals, with or without comorbidities, are more prone to develop more severe cases of COVID-19, including those related to blood perturbations, we investigated the whole blood transcriptome data during aging using the Genotype-Tissue Expression (GTEx) database [14, 15]. This strategy provided significant insights into age-associated target genes and how they can predict SARS-CoV-2 interactions in aged blood components. Furthermore, we evaluated the involvement of T cell exhaustion-associated regulatory genes and type I interferon-regulated genes, which are altered in severe COVID-19, using the data obtained from whole blood during aging.
Materials and methods
Whole blood transcriptome during aging
We used whole blood RNA-seq data from 670 males and females available at the GTEx portal (release V8) (https://www.gtexportal.org/) [16]. The BioJupies platform (https://amp.pharm.mssm.edu/biojupies/) [17] was used to find the differentially expressed genes (DEGs) in whole blood samples over aging (20–79 years old). The samples were selected and matched per age range: 30–39, 40–49, 50–59, 60–69, and 70–79. Then, age ranges were individually compared with young adults (aged 20–29). Genes with log2 fold-change ≥|1| ≤|-1| and false discovery rate (FDR) < 0.05 were considered as DEGs (Supplementary Tables 1–5). The DEGs were used to identify protein–protein interaction networks and perform gene ontology enrichment analyses.
Virus-host PPIs overlapping DEGs in whole blood with SARS-CoV-related perturbations
We first selected potential SARS-CoV-2 mediators using the human proteins available in the COVID-19 Cell Atlas (https://www.covid19cellatlas.org/) [18, 19]. These genes included SARS-CoV-2 entry receptor (ACE2), entry-associated proteases (TMPRSS2, CTSB, and CTSL), cathepsins (CTSA, CTSC, CTSD, CTSE, CTSF, CTSG, CTSH, CTSK, CTSO, CTSS, CTSV, CTSW, and CTSZ), and receptor-associated enzymes (ANPEP, DPP4, ST6GAL1, and ST3GAL4). In addition to this screening, we compared upregulated and downregulated DEGs in the whole blood during aging with the corresponding proteins that interact with human coronaviruses (HCoVs) (Table S9) by using publicly available databases. To uncover HCoVs-human PPIs, we used Pathogen–Host Interactome Prediction data using Structure Similarity (P-HIPSTer, http://phipster.org/) database. P-HIPSTer is a broad protein catalog of the virus-human interactions upon structural information with an experimental validation rate of approximately 76% [12]. Although this interactome is a good predictor of different SARS-CoV strains, our research has used P-HIPSTer to identify proteins potentially interacting with SARS-CoV-2 in other functional tissues [20, 21]. More recently, a multi-omics study revealed significant cellular interactions related to the perturbations of SARS-CoV-2 and SARS-CoV at different levels [22].
The relative expression of FASLG, CTSW, CTSE, VCAM1, and BAG3 (TMM normalized; V8 cohort) was also performed independently in male and female samples using one-way ANOVA followed by Tukey’s test. The results were analyzed with GraphPad Prism v. 6.00 for Windows (GraphPad Software, La Jolla, California, USA). Significant differences were set at P < 0.05.
Gene ontology enrichment analysis of differentially expressed genes during aging
We performed the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis and gene ontology enrichment analysis (biological processes) to identify the functions of whole blood-associated DEGs in each age range by using the EnrichR database (http://amp.pharm.mssm.edu/Enrichr/) [23]. Top enriched terms were generated according to the lowest P-value < 0.05 (Fisher’s exact test). The molecular function and protein class related to the blood components during aging were performed in the PANTHER classification system v. 11.0 (http://www.pantherdb.org/) [24]. We used the UniProtKB database (http://www.uniprot.org/) to obtain functional information of the identified proteins.
PPI networks based on blood gene expression profile during aging
The genes that appeared overexpressed in aged blood samples were analyzed by STRING online tool (https://string-db.org/) [25]. The metasearch STRING database (Search Tool for Retrieval of Interacting Genes, v. 10.5) was used for mapping PPI enrichment. We considered the following settings: text mining, databases, experiments, and co-occurrence as sources of interaction. The minimum interaction score was 0.900 (highest confidence); in the networks, the disconnected nodes were hidden to show reliable interactions exclusively. The PPI enrichment P-value indicates the statistical significance registered by STRING (Accessed in October 2020).
Single-cell transcriptomic analysis of human peripheral blood cells and bronchoalveolar fluid
We investigated the expression of selected genes (FASLG, CTSW, CTSE, VCAM, and BAG3) in distinct blood cell populations based on previously published human single-cell RNA-seq data [26]. This single-cell dataset is available at the COVID-19 Cell Atlas (https://www.covid19cellatlas.org/) and was explored using the R package Seurat v. 4.0.3 [27]. The dataset includes peripheral blood mononuclear cells (PBMCs) from severe patients hospitalized with COVID-19 (n = 7), patients with acute respiratory distress syndrome (n = 4), and healthy controls displaying no disease (n = 6). In addition, we also investigate single-cell gene expression using immune cells present in the bronchoalveolar fluid (BALF) obtained from patients at moderate (n = 3) and severe/critical (n = 6) cases of COVID-19 [28].
RNA-sequencing profiles in nasopharyngeal samples of SARS-CoV-2-infected patients
According to a previously published dataset (GSE 152075) by Lieberman et al. [29], which evaluated the status of SARS-CoV-2 infection, viral load, age, and sex differences using RNA-sequencing profiles of nasopharyngeal swabs (430 PCR-confirmed SARS-CoV-2 patients vs. 54 negative controls), we reanalyzed the data to examine host responses based on the expression level of FASLG gene in female and male samples displaying low and high viral load.
T cell exhaustion and type I interferon signaling associated gene profile in the whole blood
We sought to identify the pattern of T cell exhaustion in aging using a list of 64 genes by De Biasi et al. [30]. The nine exhaustion-associated DEGs according to age were plotted in a heatmap using the webtool Morpheus (https://software.broadinstitute.org/morpheus/) [31]. We used GraphPad Prism v.9.2.0 to generate the relative expression for each exhaustion gene over aging. Next, we investigated the expression of these genes in PBMCs of COVID-19 patients [17 COVID-19 subjects and 17 healthy controls (GSE152418)] [32]. Read counts were transformed (log2 count per million or CPM), and differentially expressed transcripts between groups were identified through the webtool NetworkAnalyst 3.0 (https://www.networkanalyst.ca/) [33] using DESeq2 pipeline. We applied the statistical cut-offs of |log2 fold-change|> 1 and adjusted P-value < 0.05 to determine DEGs.
We further searched the expression pattern of type I interferon (IFN)-associated genes based on the list of DEGs. First, the identification of type I IFN-associated genes was performed with Interferome V2.01 (http://www.interferome.org/interferome/home.jspx) [34]. The up- and downregulated genes by age were displayed using CIRCOS Plot (http://circos.ca/) [35]. The type I IFN-associated genes during aging were analyzed by Spearman’s correlation and represented in a similarity matrix.
Data representation and analysis
The comparison of blood candidate genes was based on Venn diagrams using the Venny 2.0 tool (https://bioinfogp.cnb.csic.es/tools/venny/index.html) [36]. Heatmaps and scatter plots for clustering analyses were performed using the webtool Morpheus (https://software.broadinstitute.org/morpheus) [31]. Metascape was used to provide GO terms enrichment [37] obtained from aged blood genes that potentially interact with SARS-CoV-2. One-way ANOVA complemented by Tukey’s test was used to compare the age range by each gene. Gene clustering was normalized and analyzed by k-means. Statistical analyses were performed using R software. The statistical cut-off values were P < 0.05, FDR < 0.05, log2 FC ≥|1| ≤|-1|.
Results
The number of differentially expressed genes and the complexity of associated functions increase in the whole blood during aging
The number of DEGs increases with age (log2 FC ≥|1| ≤|-1| and FDR < 0.05; Tables S1–S5). As depicted in Fig. 1A, we detected an increased number of DEGs in subjects with 50–59 years old (62 DEGs; 53 up- and nine downregulated, respectively), 60–69 (251 DEGs; 212 up- and 39 downregulated, respectively), and being the older individuals of 70–79 with the highest number (913 DEGs; 498 up- and 415 downregulated, respectively). These DEGs were predicted into PPI networks associated with immune response, cytokine and receptor activity, defense response, response to a stimulus, and signaling receptor binding. Forty-one upregulated genes shared the transcriptional profile during aging relative to the 20–29 years group and are associated with cytokine-cytokine receptor interaction, lymphocyte and natural killer chemotaxis, and eosinophil migration (FDR < 0.05, combined score > 86.06; Fig. 1B, C and Tables S6 and S7). The most prominent molecular functions were binding, regulation, and catalytic activity, while protein classes included those related to immunity, transcriptional regulator, signaling molecule, and protein modifying enzyme (Fig. 1D). Seven downregulated genes shared the transcriptional profile during aging and were mainly associated with apolipoprotein receptor binding, regulation of low-density lipoprotein, protein autoprocessing, negative regulation of receptor binding, and receptor-mediated endocytosis (FDR < 0.05, combined score > 75.90; Fig. 1E, F and Tables S6–S8). The catalytic activity, molecular regulator, transducer, and transcription activities were the most common molecular functions, while protein classes included transcriptional regulator, protein binding activity, protein modifying enzyme, and a carrier protein (Fig. 1G). Upregulated and downregulated profiles of DEGs are depicted in the volcano plots (Fig. 1H).
Differentially expressed genes (DEGs) in the whole blood distributed by age. A Up- and downregulated DEGs are increased with age relative to younger individuals. B Venn diagram combining blood upregulated transcripts over the age of 50. C Top functional terms ranking for GO and KEGG pathway categories associated with upregulated shared targets in aged blood identified by gene set enrichment analysis tool EnrichR (P-value ≥ 0.020, combined score ≥ 86.06). D Pie charts depicting the distribution percentage of molecular functions and protein classes among the blood upregulated molecules (PANTHER classification). E Venn diagram combining blood downregulated transcripts over the age of 50. F Top functional terms using GO and KEGG pathway categories associated with downregulated shared targets in aged blood identified by gene set enrichment analysis tool EnrichR (P-value ≥ 0.041, combined score ≥ 75.90). G Pie charts depicting the distribution percentage of molecular functions and protein classes among the blood downregulated molecules (PANTHER classification). H Volcano plots showing blood transcriptional gene deregulation in GTEx samples during aging, represented as − log10 (adjusted P-value) and log2 fold-change differences. Dashed lines indicate cut-offs (LogFC ≥|1| ≤|− 1| and FDR < 0.05). Blue dots = downregulated targets. Red dots = upregulated targets
Virus-host PPI interactions reveal increased expression of potential targets in whole blood during aging
We used the list of DEGs during aging to identify transcripts translated into proteins that potentially interact with SARS-CoV-2 proteins based on P-HIPSTer and COVID-19 Cell Atlas databases. This analysis identified 22 genes with an age-dependent expression profile (Table S9). As depicted in Figure S1, younger individuals (20–29, 30–39, and 40–49 years old) showed similarities and were distinctly clustered than older individuals (50–59, 60–69, and 70–79 years old) considering the global gene expression profile. By comparing clusters two by two, the age range of 20–29 showed a unique mean expression pattern compared to other aging groups (Figure S2). Among DEGs translated into predicted proteins interacting with SARS-CoV1, most of them appeared overexpressed in the age range of 50–79 years old (Fig. 2A and B), while FASLG appeared overexpressed in individuals aged 50–59, 60–69, and 70–79, the CTSW, CTSE, VCAM1, and BAG3 targets were commonly overexpressed in the two older groups of individuals (60–69 and 70–79 years old). The exclusive DEGs in 60–79 years old individuals were HBQ1, HSPA5, EPB41L3, SRC, PDCD1, CD7, CD8A, IGSF9, EPB41L4A, FRMPD3, and SCN2B.
Identification of molecular targets that potentially interact with SARS-CoV-2 and its functions associated with whole blood during aging. A Venn diagram showing common and exclusive DEGs during aging after matching with SARS-CoV-2-interacting proteins. B Heat-scatter plot presenting 22 DEGs in the whole blood during aging. The color of the circles in the plot reveals the log2 FC, while the size reflects the − log10 transformed FDR adjusted P-value. Fold-change was used to represent gene expression in blood samples in the three most advanced ages (50–59, 60–69, and 70–79 years old). C PPI interaction of proteins identified as differentially regulated illustrating the top three enriched terms and pathways. Functional interaction analysis was performed with STRING (PPI enrichment P-value < 1.0e-16; minimum confidence score: 0.9). D Functional enrichment analysis (GO terms) using all DEGs of the blood during the aging process was generated in the Metascape tool (https://metascape.org)
We generated a PPI network for all over-represented targets using the STRING database (enrichment P-value < 1.0e-16, highest confidence of 0.9; Fig. 2C). This analysis highlighted FASLG, SRC, VCAM1, BAG3, and HSPA5 as factors involved in immune response, inflammation, and platelet activation and aggregation; these proteins are directly or indirectly associated with the plasma membrane. We performed gene ontology (GO) analysis using these targets to identify top enriched terms to unravel aged blood’s functional significance with virus-host PPI. The most relevant processes and molecular functions included the immune system process, cellular component organization, biological adhesion, signaling and response to a stimulus, regulation of the biological process, and developmental process (Fig. 2D). Although CTSW and CTSE proteins are related to viral endosomal escape, FASLG, VCAM1, and BAG3 interact with viral Orf8 and protein sars7a (Tables S10 and S11). Also, VCAM1 and BAG3 showed potential interaction with spike glycoprotein, E2 glycoprotein precursors, excised polyprotein 1.4369 (gene: orf1ab), and Full_polyprotein 1.4382.
Gender-dependent transcriptional responses in whole blood during aging
We further compared mean gene expression in a cohort with female and male samples to identify gender-dependent responses (GTEx, release V8). When we compared male and female blood samples together, we found that expression of FASLG, CTSW, CTSE, and BAG3 substantially increased around the age of 50 and VCAM1 around the age of 60 years (Fig. 3). The gene expression profile in the whole blood of male and female samples was similar. Notably, FASLG showed an increased expression over the age of 50 in the blood of males and females (Fig. 4A, B). Among all DEGs, CTSW presented a similar expression profile in males and females. The CTSE was higher in males > 50 years old compared to females. Notably, increased expression of VCAM1 and BAG3 was especially pronounced in males aged 60–69 (Fig. 4A, B).
Gene expression levels (TMM) of common targets in whole blood over aging. Data are represented in box plot by mean ± SD. *P < 0.05, **P < 0.001, and ***P < 0.0001 vs. young adult individuals (20–29 years old). ANOVA complemented by Tukey’s test. FASLG, tumor necrosis factor ligand superfamily member 6; CTSE, cathepsin E; CTSW, cathepsin W; VCAM1, vascular cell adhesion molecule 1; BAG3, BAG family molecular chaperone regulator 3. The number of samples per age was as follows: n = 34 (20–29 years old and 30–39 years old), n = 72 (40–49 years old), n = 130 (50–59), n = 132 (60–69), and n = 5 (70–79)
Expression of the five genes in the whole blood over aging. Gene expression levels (TMM) detected in female (A) and male (B) samples across aging. Data are represented in box plot by mean ± SD. *P < 0.05 and **P < 0.001 vs. young adult individuals (20–29 years old). ANOVA complemented by Tukey’s test. FASLG, tumor necrosis factor ligand superfamily member 6; CTSE, cathepsin E; CTSW, cathepsin W; VCAM1, vascular cell adhesion molecule 1; BAG3, BAG family molecular chaperone regulator 3
To further examine whether FASLG expression is significantly affected in SARS-CoV-2-infected patients, we compared low and high viral load samples vs. negative control samples. While low viral load patients had no significant expression level compared to their control patients, a significant overexpression of FALSG was observed in high viral load patients (> twofold higher; P = 3.38E-12) compared with negative controls (Fig. 5A). FASLG expression was also investigated in SARS-CoV-2-infected female and male samples to discriminate sex-dependent risk. SARS-CoV-2-infected females presented higher expression of FASLG (P = 0.005, Fig. 5B) compared to their negative controls, and notably, SARS-CoV-2-infected males showed even higher expression (P = 8.37E-06, Fig. 5C) compared to their negative controls. These results reinforce the role of FASLG mainly in male patients since the mortality rate and incidence of severe cases are higher in this group.
RNA-sequencing data profile of SARS-CoV-2-infected patients based on GSE 152075 dataset. A Box plot of normalized expression (log2 CPM) of FASLG represented by mean ± SD in negative control samples (n = 54 patients), low viral load (n = 99 patients), and high viral load (n = 110 patients). ANOVA complemented by Tukey’s test. B Box plot of normalized expression (log2 CPM) of FASLG represented by mean ± SD in SARS-CoV-2-infected females (n = 201 patients) compared to SARS-CoV-2-negative females (n = 30 patients). C Box plot of normalized expression (log2 CPM) of FASLG represented by mean ± SD in SARS-CoV-2-infected males (n = 176 patients) compared to SARS-CoV-2-negative males (n = 24 patients). *P < 0.05
FASL is predominantly expressed by CD8 + T and NK cells
We analyzed the expression profile of our candidate genes (FASLG, CTSW, CTSE, VCAM1, and BAG3) in single-cell RNA-sequencing (scRNA-seq) data of PBMCs (GSE150728 dataset) and BALF-associated immune cells of SARS-CoV-2-infected patients (GSE145926 dataset) to investigate their expressions in specific blood cell population. In PBMCs, CD8 + T and natural killer (NK) cells were associated with the expressions of FASLG and CTSW genes, the latter being expressed by a higher number of immune cells (Fig. 6A, B). Also, the BAG3 gene was mainly expressed in CD4 + alpha–beta memory T cells, naïve T cells, DCs, and CD14 + monocytes (Fig. 6A, B). Regarding the BALF analysis, while FASLG was expressed in NK and T cells, the CTSW gene was mainly expressed in NK, DCs, mast cells, and B and T cells. The BAG3 gene showed high expression in epithelial cells, neutrophils, macrophages, and in plasma (Fig. 6C, D). The expression of FASLG, CTSW, and BAG3 was increased in immune cells in moderate and severe/critical cases of COVID-19 (Fig. 6E). We also noticed that VCAM1 and CTSE genes were minimally expressed by PBMCs and BALF.
Single-cell gene expression analyses of FASLG, CTSE, CTSW, VCAM1, and BAG3 in peripheral blood mononuclear cells (PBMCs) and in the bronchoalveolar fluid (BALF) of patients with COVID-19. A Regional clustering depicting gene expression in distinct cell populations (PBMC) identified in the samples of SARS-CoV-2-infected patients using an UMAP. Blue dots represent individual gene expression in single cells from PBMC samples, and the range represents the minimum and maximum expression. B Bar plot showing the percentage of cells (PBMC) individually expressing the genes. C Regional clustering depicting distinct cell populations identified in the BALF samples of SARS-CoV-2-infected patients. D Bar plot showing the percentage of cells (BALF) individually expressing the genes. E Gene expression profile detected in BALF of patients with distinct cases of COVID-19. Healthy control (HC), moderate (O), severe/critical (S/C)
Gene expression profile associated with T cell exhaustion and type I IFN signaling is affected by blood aging
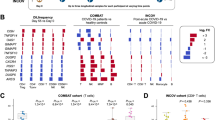
Since severity of COVID-19 is associated with T cell exhaustion, we also mined the aging blood cell transcriptome for related markers [30]. We observed four distinct gene clusters varying with blood aging. Although CCL4L2 and DUSP4 genes increased over blood aging, the LAG3, PDCD1, TIGIT, VCAM1, HLA-DRA, and TOX increased until 60–69 years old, but decreased in the last age range of 70–79. Notably, the gene RGS2, a regulator of the G-protein signaling 2, was considerably reduced in the whole blood of aged individuals compared to younger ages (Fig. 7A and B). By explicitly examining the gene profile associated with exhaustion of T cells in healthy patients and patients with different COVID-19 modalities (moderate, severe, and intensive care unit cases; GSE152418 dataset), we identified gene clusters potentially involved with immune cell exhaustion. Of note, RGS2 and HLA-DRA genes showed a more explicit expression level reduction regarding the severity of COVID-19 (Fig. 7C and D).
Analysis of gene profile associated with T cell exhaustion and type I IFN signaling in the whole blood of aging patients. A Heatmap showing blood gene variation in individuals with different ages. B Relative expression of T cell exhaustion-associated genes during aging. C Heatmap illustrating exhaustion-related gene expression in patients with different modalities of COVID-19. D Relative mRNA expression of RGS2 in healthy subjects and in moderate, severe, and intensive care unit (ICU) COVID-19 patients. E Heatmap of the genes associated with type I IFN response by patients’ ages. Two distinct patterns of clusterization are plotted showing gene subsets that decrease and increase with aging. Data were normalized by TPM + 1 and visualized as high and low values
Based on the IFN-regulated genes, we further evaluated the expression of a set of genes associated with type I IFN signaling in the blood aging. Initially, we detected two distinct gene clusters following the blood age ranges (Fig. 7E). Notably, the number of IFN-associated genes increased in the age range of 60–69 and 70–79 years old (Table S12). Specifically at the age of 70–79 years old, we detected 19 genes (ALB, CRP, LBP, CREB3L3, MFSD2A, FTCD, AKR1D1, CDKN1C, SLC38A3, JUN, ATF3, PLIN2, CTH, CYP4A11, CYP4A22, PAH, SAA1, SAA2, and PHLDA2) with increased expression over blood aging whereas 18 genes (CD1E, CD274, BATF2, CYP1A1, SLC5A9, AIM2, GCA, NMI, S100A8, S100A12, HGF, ANKRD22, APBB2, TNFSF10, SPTLC2, SAMD9, MNDA, and LGSN) had reduced expression (Fig. 7E, Figure S3). We also examined the components needed for CoV-2 sensing, clearance, and recognition, but no conclusive change was observed with blood aging (Figure S4).
Collectively, Fig. 8 highlights the possible mechanistic role whereby the alteration in the expressions of these genes during blood aging might predispose to SARS-CoV-2 dissemination and disease complication. In addition, these alterations may result in blood components dysfunctions that potentially interfere with inflammation and coagulation while inducing T cell exhaustion and defective IFN signaling.
Blood differences between older and younger individuals that might predispose to severe cases of COVID-19. While the impact of circulating SARS-CoV-2 or viral particles is associated with reduced secretion of pro-inflammatory cytokines in younger adults, a hyperinflammatory cascade accompanied by lymphocytopenia, T cell exhaustion, and possibility of blood clots formation is frequently observed in older individuals. HBQ1, hemoglobin subunit theta 1; HSPA5, heat shock protein family A (Hsp70) member 5; EPB41L3, erythrocyte membrane protein band 4.1 like 3; SRC, SRC proto-oncogene; PDCD1, programmed cell death protein 1; FRMPD3, FERM and PDZ domain containing 3; CD8A, CD8 alpha chain; CD7, member of the immunoglobulin superfamily; IGSF9, immunoglobulin superfamily member 9; EPB41L4A, erythrocyte membrane protein band 4.1 like 4A; FASLG, tumor necrosis factor ligand superfamily member 6; CTSE, cathepsin E; CTSW, cathepsin W; VCAM1, vascular cell adhesion molecule 1; BAG3, BAG family molecular chaperone regulator 3; CCL4L2, C–C motif chemokine ligand 4 like 2; DUSP4, dual specificity phosphatase 4; LAG3, lymphocyte activating 3; PDCD1, programmed cell death protein 1; TIGIT, T cell immunoreceptor with Ig and ITIM domains; HLA-DRA, major histocompatibility complex, class II, DR alpha; TOX, thymocyte selection-associated high mobility group box; RGS2, regulator of G protein signaling 2; IFNs, interferons; ?, uncertain mechanisms
Discussion
The multisystemic involvement associated with rapid clinical deterioration is among the hallmarks of COVID-19-related mortality. Although there is no evidence of transmission and dissemination of viral particles via the bloodstream [38], blood components might be involved in virus dissemination and disease aggressiveness; recent approaches uncovered that coronavirus’s immune responses persist beyond 6 months [39]. To better understand what happens with the blood of patients over aging, we investigated expression profiles of host factors predicted to interact with SARS-CoV-2 components using whole blood samples of a young adult compared with older adult individuals. In general, a distinct profile of blood targets was clustered with younger ages (20–49) and older ages (50–79), in which the most distant groups (20–29 years old versus 70–79 years old) displayed an inverse gene expression pattern. The most pronounced effects were observed over the age of 50 and included higher expression of SARS-CoV-2-related genes (e.g., genes involved in immune response, inflammation, cell component and adhesion, biological process, and platelet activation/aggregation). The increase in inflammatory mediators strongly correlates with disease severity within the conception of cytokine storm [40, 41]. In contrast to early infection, the advanced disease caused by SARS-CoV is associated with low levels of the antiviral IFNs and high levels of interleukin (IL)-1β, IL-6, and TNF and chemokines (CCL-2, CCL-3, and CCL-5) secreted by coronavirus-infected immune cells [42, 43]. Despite considerable efforts elucidating the role of SARS-CoV-2 in the immune response, there are conflicting results on blood cell infection and dissemination. Song et al. [44] reported little or even absence of ACE2 expression on T cells, Tregs, Th17, NK cells, monocytes, dendritic cells, and granulocytes, in contrast to increased expression of ACE2 on tissue-related macrophages. Conversely, immune cells (e.g., monocytes, CD8 + and CD4 + T cells, and B cells) displayed a susceptibility to SARS-CoV-2 infection in in vitro studies and also in ex vivo analysis of PBMCs from patients with severe COVID-19 [45]; it is still unclear whether the virus uses alternative mechanisms of cell entry and whether these infected cells contribute to the viral spread. By examining lung tissues of COVID-19 patients, these authors further confirmed the presence of infected immune cells.
We provided herein a transcriptomic investigation to identify possible age-associated gene expression signatures. While FASLG was found to be overexpressed in the three highest age ranges of 50–59, 60–69, and 70–79 years, some cathepsins (CTSW and CTSE), adhesion-related molecule (VCAM1), and chaperone regulator molecule (BAG3) were commonly increased after the age of 60. What should be considered is that the age-dependent changes in gene expression might not necessarily constitute a regulatory feature of aging blood cells (cell subsets) but rather reflect the variable cellular composition of aged blood [46]. This may be the reason why some blood molecules (e.g., CD8A and CD7) are detected as DEG in the 60–79 years old.
FasL is a type II transmembrane and homotrimeric protein belonging to the TNF family. After binding to its Fas receptor, a type I transmembrane TNF receptor, FasL triggers apoptotic and highly inflammatory activities [47]. Although Fas-FasL signaling has shown involvement in apoptosis of immune cells and virus-infected-target cells [48], emerging evidence highlights the apoptosis-independent role of Fas-FasL on the induction of active pro-inflammatory signals in severe pathological conditions (e.g., viral infection) [49, 50]. Furthermore, FasL promotes T cell activation in humans by recruiting cFLIP to the DISC, thereby activating NF-κB and ERK/AP-1 transcription factors [51]. These activations were responsible for the secretion of IL-2 and T cell proliferation; IL-8 was also associated with NF-κB transactivation in bronchiolar epithelial cells, whereas macrophages secreted TNF-α after Fas ligation [52] without triggering apoptotic signaling. Consistent with these findings, we detected by single-cell analysis that FASL is mainly expressed by CD8 + T and NK cells in PBMCs and BALF of SARS-CoV-2-infected patients.
A recent study by Sorbera et al. [53] on specific SARS-CoV-2-induced targets reported Fas-FasL signaling involved with endothelial function and neutrophil lifespan and related SARS-CoV-2-induced apoptosis with potential viral replication. Since FASL is a signature gene for NK and CD8 + T cell-mediated cytotoxicity (which is also shown for diverse cathepsins), elevated levels during infection most likely reflect activation and/or ongoing effector functions of these cells. It is also noteworthy that the membrane-bound form of FASL is associated with inflammation [54], but soluble FASL is also related to chronic inflammation and autoimmunity. Therefore, opposing forms of FASL need to be evaluated in aged blood of COVID-19 patients at both cellular and plasma levels to make a relevant correlation with the clinical phenotype of the disease. Severe COVID-19 has been associated with hallmarks of extrafollicular B cell activation described as systemic manifestations of autoimmunity [55]. In this context, overexpression of membrane-bound FASL can exacerbate autoimmune response while promoting inflammation and renal pathology [56]. Recently, CD4 + and CD8 + effector memory and central memory T cells from COVID-19 patients (age median 37 years) were associated with FasL secretion in addition to CD25, PD-1 expression, and IFN-γ, IL-6, granzyme, and granulysin [57]. These immune responses are thought to drive antiviral immunity against SARS-CoV-2. By contrast, recent studies using blood cells of patients with severe COVID-19 found decreased levels of sFASLG and increased blood sVCAM1 levels following disease progression [58, 59]. These contradictory results on FASLG levels might be due to the kind of secreted FASLG or the particular biological response of patients unrelated to blood aging.
A comprehensive study using whole blood samples from 54 COVID-19 patients documented a dramatic increase in immature neutrophils in parallel with a decrease in CD8 + T and VD2 γδ T cells count, which is likely due to its differentiation and activation [60]. Based on this fact, we believe that the low count of FasL-associated CD8 + T cells could result from its activation. Although the role of the Fas/FasL system in lymphocytes is unclear, patients suffering from SARS-CoV-2 infection and COVID-19 disease present T cell dysfunction and a diminished number of T and NK cells in peripheral blood [61]. The immune responses to SARS-CoV-2 infection are often characterized by hyperactivation of CD4 + and CD8 + T cells [62, 63] and macrophages [64], which produce massive levels of pro-inflammatory cytokines. Current evidence reports that critically ill patients have elevated IL-6 levels compared to moderately ill patients [41]; in addition to the infiltration of inflammatory cells, immune cells have been found in patients’ lung tissues. Notably, disease-associated transcriptional change in aged whole blood had a more pronounced overlap with control blood in comparison to lung tissue transcriptome [65]. By interacting host genes with SARS-CoV-2 and blood transcriptome, Bhattacharyya and Thelma [65] further suggested that viral infection only alters expression profile already dysregulated in the elderly, thereby resulting in poor prognosis; these altered blood genes may reinforce the appearance of severe clinical manifestations including strokes, blood clots, and heart failures.
The expression of CTSW, CTSE, VCAM1, and BAG3 was further shared in the two last age ranges compared to young individuals. Cathepsins SW (CTSW) and SE (CTSE) are papain-like cysteine protease and intracellular aspartic protease, respectively. These molecules were mainly overexpressed in males compared to females. The cathepsins B/L have been described to mediate viral entry into host cells via the endosomal pathway, participating in cell death, protein degradation, autophagy, and immune activities [66,67,68]. Although CTSB and CTSL are mainly associated with SARS-CoV-2 infection, CTSW is involved in escaping viral particles from late endosomes during influenza A virus (IAV) replication [69]. Otherwise, CTSE is expressed in immune cells implicated in antigen processing MHC class II pathway [70]. Thus, targeting CTSW and CTSE may also be a promising alternative to treat COVID-19.
Aging and vascular-immune cell interactions represent an essential aspect to be considered at the endothelial level. While soluble forms of VCAM1 are often increased in the plasma of aged humans and mice, the unique nature of plasma per se may also influence the elevation of VCAM1 in younger ages [71]. Since aging is a complex factor, other parameters should be evaluated to distinguish between aged and non-aged blood of SARS-CoV-2-infected patients. More importantly, serum levels of VCAM1 are elevated in mild COVID-19 and highly increased in severe cases [72]. There is several pathological evidence of thromboembolism, diffuse endothelial inflammation, and viral infection of endothelial cells [73, 74] related to disease severity and dysfunctional coagulation. Viral RNA load in plasma and molecular changes related to endothelial dysfunction (e.g., VCAM1, angiopoietin-2, and ICAM-1) in addition to coagulation mediators suggest a potential regulatory mechanism in the pathogenesis of endotheliitis and thrombosis in COVID-19 [75]. Whether VCAM1 expression is varying in circulating inflamed endothelial cells or in hematopoietic/non-hematopoietic cells to mediate cell-to-cell contact and cell migration needs further elucidation. Importantly, it is of great value to investigate the expression of cell adhesion molecules in aged blood of COVID-19 patients.
The BAG3, an anti-apoptotic co-chaperone molecule referred to as BCL2-associated athanogene 3, was upregulated in naive T cells, CD4 + T cells, and CD14 + T cells from aged individuals. BAG3 is involved in various biological processes such as cell survival and apoptosis, cellular stress response, and cell migration, and is suggested to be part of the SARS-CoV machinery for replication [76]. In this context, BAG3 inhibition seems to promote a significant suppression of viral replication. Like VCAM1, the BAG3 is predicted to interact with spike (S) glycoproteins. These surface molecules favor virus attachment, fusion, and entry into host cells as a direct target involved in immune responses, being lately evaluated for design and development of the S protein-based vaccines [77]. After SARS‐CoV‐2 enters the bloodstream, a cascade of events occurs resulting in blood clots and strokes [78]. We verified upregulation of proto-oncogene (SRC) and heat shock protein 70 member 5 (HSPA5) genes in aged individuals (70–79 years old); these are linked to platelet aggregation and activation. The other upregulated targets showed involvement in adaptive immunity, immunoglobulin domain, T cell receptor signalings, and cell adhesion.
Although CCL4L2 and DUSP4 genes increased over blood aging, the LAG3, PDCD1, TIGIT, VCAM1, HLA-DRA, and TOX increased until 60–69 years old, but decreased in the last age range of 70–79. Most of these immune-inhibitory receptors have been associated with severe COVID-19 [79], and exhausted T cells can undergo apoptosis with a marked decline in specific T cells [80]. The expression of RGS2 was inversely associated with blood aging and severe COVID-19. Since the regulators of G protein signaling (e.g., RGS2) are linked to T cell proliferation and IL-2 production [81], its deficiency may impair antiviral immunity. The reduction in RGS2 is associated with enhanced inflammation, immune alterations, and hypertension in animal models. Rgs2-deficient mice developed airways hyperresponsiveness with increased resistance and low compliance. These bronco-inflammatory responses were accompanied by high infiltration of inflammatory cells and increased expression of CCL3, CCL11, CXCL9, and 10 [82]. Also, mice carrying the RGS2 gene mutation showed impaired antiviral immunity [83]. The expression levels of RGS2 are also inversely correlated to hypertension and heart failure [84], which is considered a sequela as well as risk factor of COVID-19. The IFN-related genes were quite compromised in individuals aged 70–79 years old. Recently, CITE-seq analysis of PBMCs showed a lack of expression of genes related to type I IFN in COVID-19 patients [32]. Based on the KEGG and GO databases, the IFN-related genes that were downregulated in advanced ages are highly involved with positive regulation of defense response and intracellular signaling (adjusted P-value = 0.007), and cellular response to IFN (adjusted P-value = 0.008). Otherwise, the upregulated IFN-related genes correlated with Toll-like receptor signaling (adjusted P-value = 0.001) and acute inflammatory response (adjusted P-value = 0.0006). This aberrant panel of IFN-stimulated genes can interfere with the effector function of immune cells. Of note, the products of the highly expressed IFN-related genes in aged blood included molecules associated with elevated mortality rate in COVID-19 patients, such as c-reactive protein and albumin [85], lipopolysaccharide binding protein, cAMP responsive element binding protein 3 like 3, and serum amyloid A1 and A2 [86]. The immune markers CD274, BATF2, and TNFSF10, which were found to be decreased in aged blood, appeared significantly increased with disease severity [87]; this may be due to an immune response triggered by the viral infection, since age itself is unable to change these gene expression levels.
Caveats and limitations of the study is that this is an observational interactive study, and facing the absence of a specific validation targeting the main components of our model, most of the results are hypothesis-generating. Despite some divergent results, we confirmed the variation of gene profile in aged blood using different COVID-19 datasets. Furthermore, considering that age represents a confounding factor in different pathologies, we cannot determine whether the blood changes are caused by the aging process itself, SARS-CoV-2 infection, or a combination of these events. While age-dependent changes in FASLG expression have already been recognized and were also linked to aging lung and respiratory disease, there are conflicting data on its active form, expression, and serum levels regarding inflammation and apoptosis [88]. Another important aspect to be disclosed is that lymphocytopenia, a common cause associated with COVID-19, could not be evaluated using the whole blood samples available from the GTEx Biobank. Despite these limitations, our dataset-based model to understand the SARS-CoV-2 pathophysiology in whole blood in elderly matched with SARS-CoV-2-infected patients with high viral load offering a predictive model that can serve as a template for future intervention design in older patients with severe disease. What is particularly novel in our study is the recognition of the normal transcriptomic profile of whole blood and how its age-dependent variation could predispose individuals to severe COVID-19 regardless of any other specific factor (e.g., comorbidities). Future approaches are needed to evaluate the role of these blood targets considering COVID-19-related comorbidities and individual physical conditions over aging.
In summary, we demonstrated increased blood gene expressions of FASLG, BAG3, VCAM1, CTSW, and CTSE in aged patients which are possibly contributors to the severe forms of COVID-19. Through single-cell analysis using COVID-19 patients, we suggested the involvement of FASLG and CTSW associated with NK and CD8 + T cells while BAG3 was associated with CD4 + T cells, naive T cells, and CD14 + monocytes. Moreover, T cell exhaustion and IFN-associated genes were affected over blood aging, thereby showing that age itself could be a predisposing factor to severe disease. Our study highlighted these molecules as potential candidates and therapeutic targets of COVID-19 since they are involved in vascular dysfunction and altered immune response during aging. Additional studies are encouraged to test the presence of these biomarkers in the blood over different disease modalities.
Data availability
Partial data generated or analyzed in this study are included in this published article. The data presented in this study are openly available in bioRxiv at https://doi.org/10.1101/2020.12.04.41249.
References
Boulware DR, Pullen MF, Bangdiwala AS, Pastick KA, Lofgren SM, Okafor EC et al (2020) A randomized trial of hydroxychloroquine as postexposure prophylaxis for COVID-19. N Engl J Med NEJMoa2016638
Dhama K, Khan S, Tiwari R, Sircar S, Bhat S, Malik YS et al (2020) Coronavirus disease 2019—COVID-19. Clin Microbiol Ver 33:e00028-e120
Liu PP, Blet A, Smyth D, Li H (2020) The science underlying COVID-19: implications for the cardiovascular system. Circulation 142:68–78
Porfidia A, Valeriani E, Pola R, Porreca E, Rutjes AWS, Di Nisio M (2020) Venous thromboembolism in patients with COVID-19: systematic review and meta-analysis. Thromb Res 196:67–74
Al-Samkari H, Karp Leaf RS, Dzik WH, Carlson JCT, Fogerty AE, Waheed A (2020) COVID-19 and coagulation: bleeding and thrombotic manifestations of SARS-CoV-2 infection. Blood 136:489–500
Coccheri S (2020) COVID-19: the crucial role of blood coagulation and fibrinolysis. Intern Emerg Med 15:1369–1373
Liu K, Chen Y, Lin R, Han K (2020) Clinical features of COVID-19 in elderly patients: a comparison with young and middle-aged patients. J Infect 80:e14–e18
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y et al (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506
Zietz M, Zucker J, Tatonetti NP (2020) Testing the association between blood type and COVID-19 infection, intubation, and death. MedRxiv. https://doi.org/10.1101/2020.04.08.20058073
Wu BB, Gu D-Z, Yu J-N, Yang J, Shen W-Q (2020) Association between ABO blood groups and COVID-19 infection, severity and demise: a systematic review and meta-analysis. Infect Genet Evol 84:104485
Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F (2020) Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov 6:14
Lasso G, Mayer SV, Winkelmann ER, Chu T, Elliot O, Patino-Galindo JA et al (2019) A structure-informed atlas of human-virus interactions. Cell 178:1526-1541.e16
Yang S, Fu C, Lian X, Dong X, Zhang Z (2019) Understanding human-virus protein-protein interactions using a human protein complex-based analysis framework. Systems 4:e00303–18
Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S et al (2013) The genotype-tissue expression (GTEx) project. Nat Genet 45:580–585
Ardlie KG, Deluca DS, Segrè AV, Sullivan TJ, Young TR, Gelfand ET et al (2015) Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348:648–660
Robinson MD, Oshlack AA (2010) Scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11:R25
Torre D, Lachmann A, Ma’ayan A (2018) BioJupies: automated generation of interactive notebooks for RNA-seq data analysis in the cloud. Cell Syst 1–6
Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W et al (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273
Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S et al (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280
Moraes D, Paiva BVB, Cury SS, Ludwig RG, Araújo Junior JP, Mori MAS, Carvalho RF (2021) Prediction of SARS-CoV interaction with host proteins during lung aging reveals a potential role for TRIB3 in COVID-19. Aging Dis 12:42–49
Constantino FB, Cury SS, Nogueira CR, Carvalho RF, Justulin LA (2020) Prediction of non-canonical routes for SARS-CoV-2 infection in human placenta cells. BioRxiv. https://doi.org/10.1101/2020.06.12.148411
Stukalov A, Girault V, Grass V, Karayel O, Bergant V, Urban C et al (2021) Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature 594:246–252
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z et al (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44:W90–W97
Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D et al (2017) PANTHER version 11: expanded annotation data from gene ontology and reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45:D183–D189
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J et al (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613
Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martínez-Colón GJ, McKechnie JL et al (2020) A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med 1070–1076
Hao Y, Hao S, Andersen-Nissen E III, WMM, Zheng S, Butler A et al (2021) Integrated analysis of multimodal single-cell data. Cell 184:3573–3587
Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J et al (2020) Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med 26:842–844
Lieberman NAP, Peddu V, Xie H, Shrestha L, Huang ML, Mears MC et al (2020) In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age. PLoS Biol 18:e3000849
De Biasi S, Meschiari M, Gibellini L, Bellinazzi C, Borella R et al (2020) Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat Commun 11:3434
Starruß J, de Back W, Brusch L, Deutsch A (2014) Morpheus: a user-friendly modeling environment for multiscale and multicellular systems biology. Bioinformatics 30:1331–1332
Arunachalam PS, Wimmers F, Ka Pun Mok C, Perera RAPM, Scott M, Hagan T et al (2020) Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 369:1210–1220
Zhou G, Soufan O, Ewald J, Hancock REW, Basu N, Xia J (2019) NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res 47:W234–W241
Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H et al (2013) Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res 41:D1040–D1046
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D et al (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645
Oliveros JC (2015) Venny. An interactive tool for comparing lists with Venn’s diagrams. https://bioinfogp.cnb.csic.es/tools/venny/index.html.
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O et al (2019) Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10:1523
Cappy P, Candotti D, Sauvage V, Lucas Q, Boizeau L, Gomez J et al (2020) No evidence of SARS-CoV-2 transfusion transmission despite RNA detection in blood donors showing symptoms after donation. Blood 136:1888–1891
COVID research updates (2020) immune responses to coronavirus persist beyond 6 months. Nature. https://doi.org/10.1038/d41586-020-00502-w
Bhaskar S, Sinha A, Banach M, Mittoo S, Weissert R, Kass JS et al (2020) Cytokine storm in COVID-19-immunopathological mechanisms, clinical considerations, and therapeutic approaches: the REPROGRAM consortium position paper. Front Immunol 11:1648
Tang Y, Liu J, Zhang D, Xu Z, Ji J, Wen C (2020) Cytokine storm in COVID-19: the current evidence and treatment strategies. Front Immunol 11:1708
Cheung CY, Poon LLM, Ng IHY, Luk W, Sia S-F, Wu MHS (2005) Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J Virol 79:7819–7826
Law HKW, Cheung CY, Ng HY, Sia SF, Chan YO, Luk W (2005) Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood 106:2366–2374
Song X, Hu W, Yu H, Zhao L, Zhao Y, Zhao X et al (2020) Little to no expression of angiotensin-converting enzyme-2 on most human peripheral blood immune cells but highly expressed on tissue macrophages. Cytometry A 1–10
Pontelli MC, Castro IA, Martins RB, Veras FP, La Serra L, Nascimento DC et al (2020) Infection of human lymphomononuclear cells by SARS-CoV-2. bioRxiv 2020.07.28.225912
Carr EJ, Dooley J, Garcia-Perez JE, Lagou V, Lee JC, Wouters C et al (2016) The cellular composition of the human immune system is shaped by age and cohabitation. Nat Immunol 17:461–468
Chan FK, Chun HJ, Zheng L, Siegel RM, Bui KL, Lenardo MJ (2000) A domain in TNF receptors that mediates ligand-independent receptor assembly and signalings. Science 288:2351–2354
Bień K, Sokołowska J, Bąska P, Nowak Z, Stankiewicz W, Krzyzowska M (2015) Fas/FasL pathway participates in regulation of antiviral and inflammatory response during mousepox infection of lungs. Mediators Inflamm 2015:1–13
Amoras ESG, Gomes STM, Freitas FB, Santana BB, Ishak G, Araújo MTF et al (2016) Intrahepatic mRNA expression of FAS, FASL, and FOXP3 genes is associated with the pathophysiology of chronic HCV infection. PLoS One 11:e0156604
Krzyzowska M, Shestakov A, Eriksson K, Chiodi F (2011) Role of Fas/FasL in regulation of inflammation in vaginal tissue during HSV-2 infection. Cell Death Dis 2:e132
Kataoka T, Budd RC, Holler N, Thome M, Martinon F, Irmler M et al (2000) Burns K., Hahne M., KennedyN., Kovacsovics M., Tschopp J., The caspase-8 FLIP promotes activation of NF-kB and Erk signaling pathways. Curr Biol 10:640–648
Hagimoto N, Kuwano K, Kawasaki M, Yoshimi M, Kanako Y, Kunitake R et al (1999) Induction of IL-8 secretion and apoptosis in bronchiolar epithelial cells by Fas ligation. Am J Respir Cell Mol Biol 21:436–445
Sorbera LA, Graul AI, Dulsa C (2020) Taking aim at a fast-moving target: targets to watch for SARS-CoV-2 and COVID-19. Drugs of the Future 45:1–6
Shudo K, Kinoshita K, Imamura R, Fan H, Hasumoto K, Tanaka M et al (2001) The membrane-bound but not the soluble form of human Fas ligand is responsible for its inflammatory activity. Eur J Immunol 31:2504–2511
Woodruff MC, Ramonell RP, Nguyen DC, Cashman KS, Saini AS, Haddad NS et al (2020) Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat Immunol 21:1506–1516
Bossaller L, Rathinam VA, Bonegio R, Chiang PI, Busto P, Wespiser AR et al (2013) Overexpression of membrane-bound Fas ligand (CD95L) exacerbates autoimmune disease and renal pathology in pristane-induced lupus. J Immunol 191:2104–2114
Tavukcuoglu E, Horzum U, Inkaya AC, Unal S, Esendagli G (2021) Functional responsiveness of memory T cells from COVID-19 patients. Cell Immunol 365:104363
Abers MS, Delmonte OM, Ricotta EE, Fintzi J, Fink DL, Almeida de Jesus AA et al (2021) An immune-based biomarker signature is associated with mortality in COVID-19 patients. JCI Insight 6:e144455
Kessel C, Vollenberg R, Masjosthusmann K, Hinze C, Wittkowski H, Debaugnies F et al (2021) Discrimination of COVID-19 from inflammation-induced cytokine storm syndromes using disease-related blood biomarkers. Arthritis Rheumatol 73:1791–1799
Carissimo G, Xu W, Kwok I, Abdad MY, Chan Y-H, Fong S-W et al (2020) Whole blood immunophenotyping uncovers immature neutrophil-to-VD2 T-cell ratio as an early marker for severe COVID-19. Nat Commun 11:5243
Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M et al (2020) Immunology of COVID-19: current state of the science. Immunity 52:910–941
Chen Z, John Wherry E (2020) T cell responses in patients with COVID-19. Nature Rev Immunol 20:529–536
Song J-W, Zhang C, Fan X, Meng F-P, Xu Z, Xia P et al (2020) Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat Commun 11:1–10
Merad M, Martin JC (2020) Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nature Rev Immunol 20:355–362
Bhattacharyya U, Thelma BK (2020) Age-related gene expression alterations by SARS-CoV-2 infection contribute to poor prognosis in elderly. J Genet 99:80
Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S et al (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280
Liu T, Luo S, Libby P, Shi G-P (2020) Cathepsin L-selective inhibitors: a potentially promising treatment for COVID-19 patients. Pharmacol Ther 213:107587
Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P (2005) Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci USA 102:11876–11881
Edinger TO, Pohl MO, Yángüez E, Stertz S (2015) Cathepsin W is required for escape of influenza A virus from late endosomes. mBio 6:e00297–15
Zaidi N, Kalbacher H (2008) Cathepsin E: a mini review. Biochem Biophys Res Commun 367:517–522
Yousef H, Czupalla CJ, Lee D, Chen MB, Burke AN, Zera KA et al (2019) Aged blood impairs hippocampal neural precursor activity and activates microglia via brain endothelial cell VCAM1. Nat Med 25:988–1000
Tong M, Jiang Y, Xia D, Xiong Y, Zheng Q, Chen F et al (2020) Elevated expression of serum endothelial cell adhesion molecules in COVID-19 patients. J Infect Dis 222:894–898
Varga Z, Flammer AJ, Steiger P, Haberecker M, Andermatt R, Zinkernagel AS et al (2020) Endothelial cell infection and endotheliitis in COVID-19. Lancet 395:1417–1418
Wichmann D, Sperhake JP, Lütgehetmann M, Steurer S, Edler C, Heinemann A et al (2020) Autopsy findings and venous thromboembolism in patients with COVID-19: a prospective cohort study. Ann Intern Med 173:268–277
Bermejo-Martin JF (2020) Viral RNA load in plasma is associated with critical illness and a dysregulated host response in COVID-19. Crit Care 24:691
Zhang L, Zhang Z-P, Zhang X-E, Lin F-S, Ge F (2010) Quantitative proteomics analysis reveals BAG3 as a potential target to suppress severe acute respiratory syndrome coronavirus replication. J Virol 84:6050–6059
Duan L, Zheng Q, Zhang H, Niu Y, Lou Y, Wang H (2020) The SARS-CoV-2 spike glycoprotein biosynthesis, structure, function, and antigenicity: implications for the design of spike-based vaccine immunogens. Front Immunol 11:576622
Janardhan V, Janardhan V, Kalousek V (2020) COVID-19 as a blood clotting disorder masquerading as a respiratory illness: a cerebrovascular perspective and therapeutic implications for stroke thrombectomy. J Neuroimaging 30:555–561
Herrmann M, Schulte S, Wildner NH, Wittner M, Brehm TT, Ramharter M et al (2020) Analysis of co-inhibitory receptor expression in COVID-19 infection compared to acute Plasmodium falciparum malaria: LAG-3 and TIM-3 correlate with T cell activation and course of disease. Front Immunol 11:1870
Saeidi A, Zandi K, Cheok YY, Saeidi H, Wong WF, Lee CYQ et al (2018) T-cell exhaustion in chronic infections: reversing the state of exhaustion and reinvigorating optimal protective immune responses. Front Immunol 9:2569
Oliveira-Dos-Santos AJ, Matsumoto G, Snow BE, Bai D, Houston FP, Whishaw IQ et al (2000) Regulation of T cell activation, anxiety, and male aggression by RGS2. Proc Natl Acad Sci USA 97:12272–12277
George T, Bell M, Chakraborty M, Siderovski DP, Giembycz MA, Newton R (2017) Protective roles for RGS2 in a mouse model of house dust mite-induced airway inflammation. PLoS One 12:e0170269
Kehrl JH, Sinnarajah S (2002) RGS2: a multifunctional regulator of G-protein signaling. Int J Biochem Cell Biol 34:432–438
Tsang S, Woo AY, Zhu W, Xiao RP (2010) Deregulation of RGS2 in cardiovascular diseases. Front Biosci 2:547–557
Bannaga AS, Tabuso M, Farrugia A, Chandrapalan S, Somal K, Lim VK et al (2020) C-reactive protein and albumin association with mortality of hospitalised SARS-CoV-2 patients: a tertiary hospital experience. Clin Med 20:463–467
Mahmud I, Garrett TJ (2020) Mass spectrometry techniques in emerging pathogens studies: COVID-19 perspectives. J Am Soc Mass Spectrom 31:2013–2024
Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N et al (2020) Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 369:718–724
Wallach-Dayan SB, Petukhov D, Ahdut-HaCohen R, Richter-Dayan M, Breuer R (2021) sFasL—the key to a riddle: immune responses in aging lung and disease. Int J Mol Sci 22:2177
Funding
We gratefully thank the following grants that supported the development of this study: São Paulo Research Foundation (FAPESP grant # 2019/00906–6 to LGAC), and the National Council for Scientific and Technological Development, CNPq (Process 304108/2020–0 to LGAC and 311530/2019–2 to RFC) and Santander Universidades (Banco Santander Brasil S/A) for COVID-19 research (to RFC).
Author information
Authors and Affiliations
Contributions
LGAC and RFC: conception of the idea, design of the study, and drafted the manuscript; JSS and PPF: statistical analysis and acquisition of data; MON and MCM: participated in data analysis and interpretation. All authors significantly contributed with compilation of the literature and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval
All data in this article were reanalyzed from previous published datasets.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
de Almeida Chuffa, L.G., Freire, P.P., dos Santos Souza, J. et al. Aging whole blood transcriptome reveals candidate genes for SARS-CoV-2-related vascular and immune alterations. J Mol Med 100, 285–301 (2022). https://doi.org/10.1007/s00109-021-02161-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-021-02161-4