Abstract
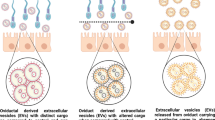
Embryo-derived extracellular vesicles (EVs) may play a role in mediating the embryo-maternal dialogue at the oviduct, potentially carrying signals reflecting embryo quality. We investigated the effects of bovine embryo-derived EVs on the gene expression of bovine oviductal epithelial cells (BOECs), and whether these effects are dependent on embryo quality. Presumptive zygotes were cultured individually in vitro in culture medium droplets until day 8 while their development was assessed at day 2, 5 and 8. Conditioned medium samples were collected at day 5 and pooled based on embryo development (good quality embryo media and degenerating embryo media). EVs were isolated from conditioned media by size exclusion chromatography and supplemented to primary BOEC monolayer cultures to evaluate the effects of embryo-derived EVs on gene expression profile of BOEC. Gene expression was quantified by RNA-seq and RT-qPCR. A total of 7 upregulated and 18 downregulated genes were detected in the BOECs supplemented with good quality embryo-derived EV compared to the control. The upregulated genes included interferon-τ-induced genes, such as OAS1Y, MX1 and ISG15, which have previously been reported as upregulated in the oviductal epithelial cells in the presence of embryos. Of the upregulated genes, OAS1Y and MX1 were validated with RT-qPCR. In contrast, only one differentially expressed gene was detected in BOECs in response to degenerating embryo-derived EVs, suggesting that oviductal responses are dependent on embryo quality. Our results support the hypothesis that embryo-derived EVs are involved in embryo-maternal communication at the oviduct and the oviductal response is dependant on the embryo quality.
Key messages
• Extracellular vesicles (EVs) released by individually cultured pre-implantation bovine embryos can alter the gene expression of primary oviductal epithelial cells.
• The oviductal response, in terms of gene expression, to the bovine embryo-derived EVs varied depending on the embryo quality.
• In vivo, the oviduct may have the ability to sense the quality of the pre-implantation embryos.
• The observed effect of embryo-derived EVs on oviductal epithelial cells could serve as a non-invasive method of evaluating the embryo quality.




Similar content being viewed by others
Data availability
The RNA sequencing data has been uploaded to the NCBI SRA repository (www.ncbi.nlm.nih.gov/sra) under NCBI BioProject ID PRJNA660346.
References
Bauersachs S, Ulbrich SE, Zakhartchenko V, Minten M, Reichenbach M, Reichenbach HD, Blum H, Spencer TE, Wolf E (2009) The endometrium responds differently to cloned versus fertilized embryos. Proc Natl Acad Sci U S A 106:5681–5686
Mansouri-Attia N, Sandra O, Aubert J, Degrelle S, Everts RE, Giraud-Delville C, Heyman Y, Galio L, Hue I, Yang X (2009) Endometrium as an early sensor of in vitro embryo manipulation technologies. Proc Natl Acad Sci U S A 106:5687–5692
Macklon NS, Brosens JJ (2014) The human endometrium as a sensor of embryo quality. Biol Reprod 91:98
Coy P, García-Vázquez FA, Visconti PE, Avilés M (2012) Roles of the oviduct in mammalian fertilization. Reproduction 144:649–660
Besenfelder U, Havlicek V, Brem G (2012) Role of the oviduct in early embryo development. Reprod Domest Anim 47:156–163
Cordova A, Perreau C, Uzbekova S, Ponsart C, Locatelli Y, Mermillod P (2014) Development rate and gene expression of IVP bovine embryos cocultured with bovine oviduct epithelial cells at early or late stage of preimplantation development. Theriogenology 81:1163–1173
Hamdi M, Sánchez-Calabuig MJ, Rodríguez-Alonso B, Arnal SB, Roussi K, Sturmey R et al (2019) Gene expression and metabolic response of bovine oviduct epithelial cells to the early embryo. Reproduction 158:85–94
Schmaltz-Panneau B, Cordova A, Dhorne-Pollet S, Hennequet-Antier C, Uzbekova S, Martinot E, Doret S, Martin P, Mermillod P, Locatelli Y (2014) Early bovine embryos regulate oviduct epithelial cell gene expression during in vitro co-culture. Anim Reprod Sci 149:103–116
García EV, Hamdi M, Barrera AD, Sánchez-Calabuig MJ, Gutiérrez-Adán A, Rizos D (2017) Bovine embryo-oviduct interaction in vitro reveals an early cross talk mediated by BMP signaling. Reproduction 153:631–643
Maillo V, Gaora P, Forde N, Besenfelder U, Havlicek V, Burns GW et al (2015) Oviduct-embryo interactions in cattle: two-way traffic or a one-way street? Biol Reprod 92:144
Cruz L, Romero JAA, Iglesia RP, Lopes MH (2018) Extracellular vesicles: decoding a new language for cellular communication in early embryonic development. Front Cell Dev Biol 6. https://doi.org/10.3389/fcell.2018.00094
Royo F, Falcon-Perez JM (2012) Liver extracellular vesicles in health and disease. J Extracell Vesicles 1. https://doi.org/10.3402/jev.v1i0.18825
Hurwitz SN, Meckes DG (2019) Extracellular vesicle integrins distinguish unique cancers. Proteomes 7. https://doi.org/10.3390/proteomes7020014
Tkach M, Théry C (2016) Communication by extracellular vesicles: where we are and where we need to go. Cell 164:1226–1232
van der Pol E, Böing AN, Harrison P, Sturk A, Nieuwland R (2012) Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev 64:676–705
Lopera-Vasquez R, Hamdi M, Fernandez-Fuertes B, Maillo V, Beltran-Brena P, Calle A et al (2016) Extracellular vesicles from BOEC in in vitro embryo development and quality. PLoS One 11:e0148083
Braicu C, Tomuleasa C, Monroig P, Cucuianu A, Berindan-Neagoe I, Calin GA (2015) Exosomes as divine messengers: are they the Hermes of modern molecular oncology? Cell Death Differ 22:34–45
Kalra H, Drummen GPC, Mathivanan S (2016) Focus on extracellular vesicles: introducing the next small big thing. Int J Mol Sci 17. https://doi.org/10.3390/ijms17020170
Giacomini E, Vago R, Sanchez AM, Podini P, Zarovni N, Murdica V, Rizzo R, Bortolotti D, Candiani M, Viganò P (2017) Secretome of in vitro cultured human embryos contains extracellular vesicles that are uptaken by the maternal side. Sci Rep 7:5210
Takasugi M, Okada R, Takahashi A, Virya Chen D, Watanabe S, Hara E (2017) Small extracellular vesicles secreted from senescent cells promote cancer cell proliferation through EphA2. Nat Commun 8:15729
Saadeldin IM, Kim SJ, Bin CY, Lee BC (2014) Improvement of cloned embryos development by co-culturing with parthenotes: a possible role of exosomes/microvesicles for embryos paracrine communication. Cell Reprogram 16:223–234
Mellisho EA, Velásquez AE, Nuñez MJ, Cabezas JG, Cueto JA, Fader C, Castro FO, Rodríguez-Álvarez L (2017) Identification and characteristics of extracellular vesicles from bovine blastocysts produced in vitro. PLoS One 12:e0178306
Dissanayake K, Nõmm M, Lättekivi F, Ressaissi Y, Godakumara K, Lavrits A, Midekessa G, Viil J, Bæk R, Jørgensen MM (2020) Individually cultured bovine embryos produce extracellular vesicles that have the potential to be used as non-invasive embryo quality markers. Theriogenology 149:104–116
Qu P, Qing S, Liu R, Qin H, Wang W, Qiao F, Ge H, Liu J, Zhang Y, Cui W (2017) Effects of embryo-derived exosomes on the development of bovine cloned embryos. PLoS One 12:e0174535
Pavani KC, Hendrix A, Van Den Broeck W, Couck L, Szymanska K, Lin X et al (2019) Isolation and characterization of functionally active extracellular vesicles from culture medium conditioned by bovine embryos in vitro. Int J Mol Sci 20:38
Bauersachs S, Mermillod P, Almiñana C (2020) The oviductal extracellular vesicles’ RNA cargo regulates the bovine embryonic transcriptome. Int J Mol Sci 21. https://doi.org/10.3390/ijms21041303
Es-Haghi M, Godakumara K, Häling A, Lättekivi F, Lavrits A, Viil J, Andronowska A, Nafee T, James V, Jaakma Ü (2019) Specific trophoblast transcripts transferred by extracellular vesicles affect gene expression in endometrial epithelial cells and may have a role in embryo-maternal crosstalk. Cell Commun Signal 17. https://doi.org/10.1186/s12964-019-0448-x
Nõmm M, Porosk R, Pärn P, Kilk K, Soomets U, Kõks S, Jaakma Ü (2019) In vitro culture and non-invasive metabolic profiling of single bovine embryos. Reprod Fertil Dev 31:306–314
Bó GA, Mapletoft RJ. Evaluation and classification of bovine embryos. vol. 10. 2013.
Ireland JJ, Murphee RL, Coulson PB (1980) Accuracy of predicting stages of bovine estrous cycle by gross appearance of the corpus luteum. J Dairy Sci 63:155–160
Picelli S, Faridani OR, Björklund ÅK, Winberg G, Sagasser S, Sandberg R (2014) Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 9:171–181
Brown J, Pirrung M, Mccue LA (2017) FQC Dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 33:3137–3139
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Kim D, Paggi JM, Park C, Bennett C, Salzberg SL (2019) Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37:907–915
Liao Y, Smyth GK, Shi W (2014) FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930
Robinson MD, McCarthy DJ, Smyth GK (2009) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Yu G, Wang LG, Han Y, He QY (2012) ClusterProfiler: an R package for comparing biological themes among gene clusters. Omi A J Integr Biol 16:284–287
Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K (2017) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45:D353–D361
Team RDC. A language and environment for statistical computing. R Found Stat Comput 2018;2:https://www.R-project.org.
Wickhman H. ggplot2: elegant graphics for data analysis. 2009.
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134
Idt. Integrated DNA technologies. Release 75 2014;2008:2661. https://www.idtdna.com/site (accessed February 18, 2020).
Zeisberg M, Neilson EG (2009) Biomarkers for epithelial-mesenchymal transitions. J Clin Invest 119:1429–1437
Demmers KJ, Derecka K, Flint A (2001) Trophoblast interferon and pregnancy. Reproduction 121:41–49
Hansen TR, Austin KJ, Johnson GA (1997) Transient ubiquitin cross-reactive protein gene expression in the bovine endometrium. Endocrinology 138:5079–5082
Talukder AK, Rashid MB, Yousef MS, Kusama K, Shimizu T, Shimada M, Suarez SS, Imakawa K, Miyamoto A (2018) Oviduct epithelium induces interferon-tau in bovine day-4 embryos, which generates an anti-inflammatory response in immune cells. Sci Rep 8:7850
Passaro C, Tutt D, Mathew DJ, Sanchez JM, Browne JA, Boe-Hansen GB, Fair T, Lonergan P (2018) Blastocyst-induced changes in the bovine endometrial transcriptome. Reproduction 156:219–229
Johnson GA, Austin KJ, Van Kirk EA, Hansen TR (1998) Pregnancy and interferon-tau induce conjugation of bovine ubiquitin cross-reactive protein to cytosolic uterine proteins. Biol Reprod 58:898–904
Hicks BA, Etter SJ, Carnahan KG, Joyce MM, Assiri AA, Carling SJ, Kodali K, Johnson GA, Hansen TR, Mirando MA (2003) Expression of the uterine Mx protein in cyclic and pregnant cows, gilts, and mares. J Anim Sci 81:1552–1561
Smits K, De Coninck DIM, Van Nieuwerburgh F, Govaere J, Van Poucke M, Peelman L et al (2016) The equine embryo influences immune-related gene expression in the oviduct. Biol Reprod 94:36
Zhang D, Zhang DE (2011) Interferon-stimulated gene 15 and the protein ISGylation system. J Interf Cytokine Res 31:119–130
Pavlovic J, Zürcher T, Haller O, Staeheli P (1990) Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J Virol 64:3370–3375
Sadler AJ, Williams BRG (2008) Interferon-inducible antiviral effectors. Nat Rev Immunol 8:559–568
Castelli JAC, Hassel BA, Maran A, Paranjape J, Hewitt JA, Li XL, Hsu YT, Silverman RH, Youle RJ (1998) The role of 2’-5’ oligoadenylate-activated ribonuclease L in apoptosis. Cell Death Differ 5:313–320
Ghosh A, Sarkar SN, Rowe TM, Sen GC (2001) A specific isozyme of 2′-5′ oligoadenylate synthetase is a dual function proapoptotic protein of the Bcl-2 family. J Biol Chem 276:25447–25455
Hoen EN, Cremer T, Gallo RC, Margolis LB (2016) Extracellular vesicles and viruses: are they close relatives? Proc Natl Acad Sci U S A 113:9155–9161
Pivot-Pajot C, Varoqueaux F, de Saint BG, Bourgoin SG (2008) Munc13-4 regulates granule secretion in human neutrophils. J Immunol 180:6786–6797
Birkenfeld J, Nalbant P, Yoon SH, Bokoch GM (2008) Cellular functions of GEF-H1, a microtubule-regulated Rho-GEF: is altered GEF-H1 activity a crucial determinant of disease pathogenesis? Trends Cell Biol 18:210–219
Zhao Y, Alonso C, Ballester I, Song JH, Chang SY, Guleng B, Arihiro S, Murray PJ, Xavier R, Kobayashi KS (2012) Control of NOD2 and Rip2-dependent innate immune activation by GEF-H1. Inflamm Bowel Dis 18:603–612
Almiñana C, Heath PR, Wilkinson S, Sanchez-Osorio J, Cuello C, Parrilla I, Gil MA, Vazquez JL, Vazquez JM, Roca J (2012) Early developing pig embryos mediate their own environment in the maternal tract. PLoS One 7:e33625
Betts DH, Madan P (2008) Permanent embryo arrest: molecular and cellular concepts. Mol Hum Reprod 14:445–453
Betts DH, King WA (2001) Genetic regulation of embryo death and senescence. Theriogenology 55:171–191
Graf A, Krebs S, Heininen-Brown M, Zakhartchenko V, Blum H, Wolf E (2014) Genome activation in bovine embryos: review of the literature and new insights from RNA sequencing experiments. Anim Reprod Sci 149:46–58
Assou S, Boumela I, Haouzi D, Anahory T, Dechaud H, de Vos J, Hamamah S (2011) Dynamic changes in gene expression during human early embryo development: from fundamental aspects to clinical applications. Hum Reprod Update 17:272–290
Feng D, Zhao WL, Ye YY, Bai XC, Liu RQ, Chang LF, Zhou Q, Sui SF (2010) Cellular internalization of exosomes occurs through phagocytosis. Traffic 11(5):675–687
Costa Verdera H, Gitz-Francois JJ, Schiffelers RM, Vader P (2017) Cellular uptake of extracellular vesicles is mediated by clathrin-independent endocytosis and macropinocytosis. J Control Release 266:100–108
Acknowledgements
We thank Annika Häling for her support during the laboratory work.
Funding
This work was supported by the European Union’s Horizon 2020 research and innovation programme under grant agreement no. 668989 (TRANSGENO) and 857418 (COMBIVET); the Estonian Ministry of Education and Research (grant IUT34-16); Enterprise Estonia (grant EU48695); the European Commission Horizon 2020 research and innovation programme under grant agreement 692065 (Project WIDENLIFE) and European Commission Horizon 2020 research and innovation programme under grant agreement 692299 (Project SEARMET); Rep-Eat-H2020-MSCA-COFUND 713714.
Author information
Authors and Affiliations
Contributions
KD, AF and MN developed the concept and designed the experiments; MN and YR produced embryos in vitro; QR carried out primary BOEC culture; KD carried out EV isolation, extended BOEC culture, EV supplementation and RNA extraction; KJ and AVM carried out RNAseq library preparation and RNAseq; JV did BOEC immunofluorescence assay; FL and JO carried out RNAseq data analysis; KD and KG carried out RT-qPCR; KD wrote the manuscript with conceptual contribution from JO, FL and AF; AF, ÜJ and AS supervised KD. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Code availability
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Online Resource 1
, file format-.pdf, Primer sequences of the genes used for qPCR (PDF 194 kb)
Online Resource 2
, file format-.xls, Good embryo-derived EVs supplemented BOECs vs Control BOECs, RNAseq differential gene expression analysis (XLSX 966 kb)
Online Resource 3
, file format-.xls, Degenerating embryo-derived EVs supplemented BOECs vs Control BOECs, RNAseq differential gene expression analysis (XLSX 958 kb)
Online Resource 4
, file format-.xls, Good embryo-derived EVs supplemented BOECs vs degenerating embryo-derived EVs supplemented BOECs, RNAseq differential gene expression analysis (XLSX 959 kb)
Online Resource 5
, file format-.xls, Good_vs_control_kegg_gsea, GSEA with KEGG pathway annotations based on the results of differential expression tests (XLS 120 kb)
Online Resource 6
, file format-.xls, Good_vs_degenerating_kegg_gsea, GSEA with KEGG pathway annotations based on the results of differential expression tests (XLS 118 kb)
Rights and permissions
About this article
Cite this article
Dissanayake, K., Nõmm, M., Lättekivi, F. et al. Oviduct as a sensor of embryo quality: deciphering the extracellular vesicle (EV)-mediated embryo-maternal dialogue. J Mol Med 99, 685–697 (2021). https://doi.org/10.1007/s00109-021-02042-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-021-02042-w