Abstract
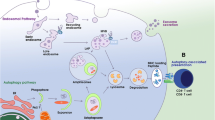
Autophagy delivers cytoplasmic constituents for lysosomal degradation. Recent studies have demonstrated that this pathway mediates resistance to pathogens and is targeted for immune evasion by viruses and bacteria. Lysosomal degradation products, including pathogenic determinants, are then surveyed by the adaptive immune system to elicit antigen-specific T cell responses. CD4+ T helper cells have been shown to recognize nuclear and cytosolic antigens via presentation by major histocompatibility complex (MHC) class II molecules after autophagy. Furthermore, some sources of natural MHC class II ligands display characteristics of autophagy substrates, and autophagosomes fuse with late endosomes, in which MHC class II loading is thought to occur. Although MHC class II antigen processing via autophagy has so far mainly been described for professional antigen-presenting cells like B cells, macrophages, and dendritic cells, it might be even more important for cells with less endocytic potential, like epithelial cells, when these express MHC class II at sites of inflammation. Therefore, autophagy might contribute to immune surveillance of intracellular pathogens via MHC class II presentation of intracellular pathogen-derived peptides.


Similar content being viewed by others
Abbreviations
- APC:
-
Antigen-presenting cell
- Atg:
-
Autophagy-related gene
- CLIP:
-
Class II-associated Ii-derived peptide
- CTL:
-
Cytotoxic T lymphocyte
- DRiP:
-
Defective ribosomal product
- EBNA1:
-
Epstein–Barr virus nuclear antigen 1
- EBV:
-
Epstein–Barr virus
- Hsc70:
-
Heat shock cognate protein 70/73
- HLA:
-
Human leukocyte antigen
- Ii:
-
Invariant chain
- LAMP:
-
Lysosomal-associated membrane protein
- MHC:
-
Major histocompatibility complex
- MIIC:
-
MHC class II-containing compartment
- Mtb:
-
Mycobacterium tuberculosis
- Th:
-
T helper cell
References
Kloetzel PM (2001) Antigen processing by the proteasome. Nat Rev Mol Cell Biol 2:179–187
Kornfeld S, Mellman I (1989) The biogenesis of lysosomes. Annu Rev Cell Biol 5:483–525
Yewdell JW, Reits E, Neefjes J (2003) Making sense of mass destruction: quantitating MHC class I antigen presentation. Nat Rev Immunol 3:952–961
Stuart LM, Ezekowitz RA (2005) Phagocytosis: elegant complexity. Immunity 22:539–550
Henell F, Berkenstam A, Ahlberg J, Glaumann H (1987) Degradation of short- and long-lived proteins in perfused liver and in isolated autophagic vacuoles-lysosomes. Exp Mol Pathol 46:1–14
Trombetta ES, Mellman I (2005) Cell biology of antigen processing in vitro and in vivo. Annu Rev Immunol 23:975–1028
De Duve C, Wattiaux R (1966) Functions of lysosomes. Annu Rev Physiol 28:435–492
Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N (2004) The role of autophagy during the early neonatal starvation period. Nature 432:1032–1036
Klionsky DJ, Emr SD (2000) Autophagy as a regulated pathway of cellular degradation. Science 290:1717–1721
Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y (2004) In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15:1101–1111
Cuervo AM, Dice JF (1996) A receptor for the selective uptake and degradation of proteins by lysosomes. Science 273:501–503
Cuervo AM, Dice JF (2000) Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci 113(Pt 24):4441–4450
Chiang HL, Terlecky SR, Plant CP, Dice JF (1989) A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science 246:382–385
Agarraberes FA, Terlecky SR, Dice JF (1997) An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J Cell Biol 137:825–834
Agarraberes FA, Dice JF (2001) Protein translocation across membranes. Biochim Biophys Acta 1513:1–24
Yoshimori T (2004) Autophagy: a regulated bulk degradation process inside cells. Biochem Biophys Res Commun 313:453–458
Klionsky DJ, Cregg JM, Dunn WA Jr, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y (2003) A unified nomenclature for yeast autophagy-related genes. Dev Cell 5:539–545
Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y (1998) A protein conjugation system essential for autophagy. Nature 395:395–398
Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19:5720–5728
Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y (2000) A ubiquitin-like system mediates protein lipidation. Nature 408:488–492
Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V (2004) Autophagy is a defense mechanism inhibiting BCG and mycobacterium tuberculosis survival in infected macrophages. Cell 119:753–766
Kim J, Klionsky DJ (2000) Autophagy, cytoplasm-to-vacuole targeting pathway, and pexophagy in yeast and mammalian cells. Annu Rev Biochem 69:303–342
Shintani T, Huang WP, Stromhaug PE, Klionsky DJ (2002) Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell 3:825–837
Suzuki K, Kamada Y, Ohsumi Y (2002) Studies of cargo delivery to the vacuole mediated by autophagosomes in Saccharomyces cerevisiae. Dev Cell 3:815–824
Simonsen A, Birkeland HC, Gillooly DJ, Mizushima N, Kuma A, Yoshimori T, Slagsvold T, Brech A, Stenmark H (2004) Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J Cell Sci 117:4239–4251
Levine B (2005) Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 120:159–162
Kirkegaard K, Taylor MP, Jackson WT (2004) Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat Rev Microbiol 2:301–314
Shintani T, Klionsky DJ (2004) Autophagy in health and disease: a double-edged sword. Science 306:990–995
Rich KA, Burkett C, Webster P (2003) Cytoplasmic bacteria can be targets for autophagy. Cell Microbiol 5:455–468
Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, Hamada S, Yoshimori T (2004) Autophagy defends cells against invading group A Streptococcus. Science 306:1037–1040
Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C (2005) Escape of intracellular shigella from autophagy. Science 307:727–731
Amer AO, Swanson MS (2005) Autophagy is an immediate macrophage response to Legionella pneumophila. Cell Microbiol 7:765–778
Talloczy Z, Jiang W, Virgin, HWt, Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B (2002) Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci U S A 99:190–195
Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B (1998) Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol 72:8586–8596
Prentice E, Jerome WG, Yoshimori T, Mizushima N, Denison MR (2004) Coronavirus replication complex formation utilizes components of cellular autophagy. J Biol Chem 279:10136–10141
Pedersen KW, van der Meer Y, Roos N, Snijder EJ (1999) Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J Virol 73:2016–2026
Schlegel A, Giddings TH Jr, Ladinsky MS, Kirkegaard K (1996) Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol 70:6576–6588
Suhy DA, Giddings TH Jr, Kirkegaard K (2000) Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J Virol 74:8953–8965
Jackson WT, Giddings TH Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K (2005) Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol 3:e156
Meyers G, Stoll D, Gunn M (1998) Insertion of a sequence encoding light chain 3 of microtubule-associated proteins 1A and 1B in a pestivirus genome: connection with virus cytopathogenicity and induction of lethal disease in cattle. J Virol 72:4139–4148
Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S (1999) SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50:213–219
Chicz RM, Urban RG, Gorga JC, Vignali DA, Lane WS, Strominger JL (1993) Specificity and promiscuity among naturally processed peptides bound to HLA-DR alleles. J Exp Med 178:27–47
Dongre AR, Kovats S, deRoos P, McCormack AL, Nakagawa T, Paharkova-Vatchkova V, Eng J, Caldwell H, Yates JR III, Rudensky AY (2001) In vivo MHC class II presentation of cytosolic proteins revealed by rapid automated tandem mass spectrometry and functional analyses. Eur J Immunol 31:1485–1494
Dengjel J, Schoor O, Fischer R, Reich M, Kraus M, Muller M, Kreymborg K, Altenberend F, Brandenburg J, Kalbacher H, Brock R, Driessen C, Rammensee HG, Stevanovic S (2005) Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci U S A 102:7922–7927
Aniento F, Roche E, Cuervo AM, Knecht E (1993) Uptake and degradation of glyceraldehyde-3-phosphate dehydrogenase by rat liver lysosomes. J Biol Chem 268:10463–10470
Fengsrud M, Raiborg C, Berg TO, Stromhaug PE, Ueno T, Erichsen ES, Seglen PO (2000) Autophagosome-associated variant isoforms of cytosolic enzymes. Biochem J 352(Pt 3):773–781
Friede T, Gnau V, Jung G, Keilholz W, Stevanovic S, Rammensee HG (1996) Natural ligand motifs of closely related HLA-DR4 molecules predict features of rheumatoid arthritis associated peptides. Biochim Biophys Acta 1316:85–101
Harris PE, Maffei A, Colovai AI, Kinne J, Tugulea S, Suciu-Foca N (1996) Predominant HLA-class II bound self-peptides of a hematopoietic progenitor cell line are derived from intracellular proteins. Blood 87:5104–5112
Rammensee HG, Bachmann J, Stevanovic S (1997) MHC ligands and peptide motifs. Springer, Berlin Heidelberg New York; Landes Bioscience, Austin
Seglen PO, Gordon PB (1982) 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A 79:1889–1892
Nimmerjahn F, Milosevic S, Behrends U, Jaffee EM, Pardoll DM, Bornkamm GW, Mautner J (2003) Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur J Immunol 33:1250–1259
Brazil MI, Weiss S, Stockinger B (1997) Excessive degradation of intracellular protein in macrophages prevents presentation in the context of major histocompatibility complex class II molecules. Eur J Immunol 27:1506–1514
Dörfel D, Appel S, Grunebach F, Weck MM, Muller MR, Heine A, Brossart P (2005) Processing and presentation of HLA class I and II epitopes by dendritic cells after transfection with in vitro transcribed MUC1 RNA. Blood 105:3199–3205
Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Münz C (2005) Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 307:593–596
Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ (1997) The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem 243:240–246
Biederbick A, Kern HF, Elsasser HP (1995) Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur J Cell Biol 66:3–14
Zhou D, Li P, Lott JM, Hislop A, Canaday DH, Brutkiewicz RR, Blum JS (2005) Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens. Immunity 22:571–581
Lich JD, Elliott JF, Blum JS (2000) Cytoplasmic processing is a prerequisite for presentation of an endogenous antigen by major histocompatibility complex class II proteins. J Exp Med 191:1513–1524
Dul JL, Davis DP, Williamson EK, Stevens FJ, Argon Y (2001) Hsp70 and antifibrillogenic peptides promote degradation and inhibit intracellular aggregation of amyloidogenic light chains. J Cell Biol 152:705–716
Barette C, Jariel-Encontre I, Piechaczyk M, Piette J (2001) Human cyclin C protein is stabilized by its associated kinase cdk8, independently of its catalytic activity. Oncogene 20:551–562
Dice JF, Goldberg AL (1975) A statistical analysis of the relationship between degradative rates and molecular weights of proteins. Arch Biochem Biophys 170:213–219
Gueguen M, Long EO (1996) Presentation of a cytosolic antigen by major histocompatibility complex class II molecules requires a long-lived form of the antigen. Proc Natl Acad Sci U S A 93:14692–14697
Blake N, Lee S, Redchenko I, Thomas W, Steven N, Leese A, Steigerwald-Mullen P, Kurilla MG, Frappier L, Rickinson A (1997) Human CD8+ T cell responses to EBV EBNA1: HLA class I presentation of the (Gly-Ala)-containing protein requires exogenous processing. Immunity 7:791–802
Levitskaya J, Sharipo A, Leonchiks A, Ciechanover A, Masucci MG (1997) Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein–Barr virus nuclear antigen 1. Proc Natl Acad Sci U S A 94:12616–12621
Lee SP, Brooks JM, Al-Jarrah H, Thomas WA, Haigh TA, Taylor GS, Humme S, Schepers A, Hammerschmidt W, Yates JL, Rickinson AB, Blake NW (2004) CD8 T cell recognition of endogenously expressed Epstein–Barr virus nuclear antigen 1. J Exp Med 199:1409–1420
Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR (2000) Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404:770–774
Reits EA, Vos JC, Gromme M, Neefjes J (2000) The major substrates for TAP in vivo are derived from newly synthesized proteins. Nature 404:774–778
Princiotta MF, Finzi D, Qian SB, Gibbs J, Schuchmann S, Buttgereit F, Bennink JR, Yewdell JW (2003) Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity 18:343–354
Ciechanover A, Finley D, Varshavsky A (1984) Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell 37:57–66
Zwart W, Griekspoor A, Kuijl C, Marsman M, van Rheenen J, Janssen H, Calafat J, van Ham M, Janssen L, van Lith M, Jalink K, Neefjes J (2005) Spatial separation of HLA-DM/HLA-DR interactions within MIIC and phagosome-induced immune escape. Immunity 22:221–233
Stromhaug PE, Berg TO, Fengsrud M, Seglen PO (1998) Purification and characterization of autophagosomes from rat hepatocytes. Biochem J 335(Pt 2):217–224
Fengsrud M, Erichsen ES, Berg TO, Raiborg C, Seglen PO (2000) Ultrastructural characterization of the delimiting membranes of isolated autophagosomes and amphisomes by freeze-fracture electron microscopy. Eur J Cell Biol 79:871–882
Lajoie P, Guay G, Dennis JW, Nabi IR (2005) The lipid composition of autophagic vacuoles regulates expression of multilamellar bodies. J Cell Sci 118:1991–2003
Berg TO, Fengsrud M, Stromhaug PE, Berg T, Seglen PO (1998) Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem 273:21883–21892
Liou W, Geuze HJ, Geelen MJ, Slot JW (1997) The autophagic and endocytic pathways converge at the nascent autophagic vacuoles. J Cell Biol 136:61–70
Kleijmeer MJ, Morkowski S, Griffith JM, Rudensky AY, Geuze HJ (1997) Major histocompatibility complex class II compartments in human and mouse B lymphoblasts represent conventional endocytic compartments. J Cell Biol 139:639–649
Starr TK, Jameson SC, Hogquist KA (2003) Positive and negative selection of T cells. Annu Rev Immunol 21:139–176
Reith W, Mach B (2001) The bare lymphocyte syndrome and the regulation of MHC expression. Annu Rev Immunol 19:331–373
Sun JC, Williams MA, Bevan MJ (2004) CD4+ T cells are required for the maintenance, not programming, of memory CD8+ T cells after acute infection. Nat Immunol 5:927–933
Bevan MJ (2004) Helping the CD8+ T-cell response. Nat Rev Immunol 4:595–602
Stalder T, Hahn S, Erb P (1994) Fas antigen is the major target molecule for CD4+ T cell-mediated cytotoxicity. J Immunol 152:1127–1133
Hahn S, Gehri R, Erb P (1995) Mechanism and biological significance of CD4-mediated cytotoxicity. Immunol Rev 146:57–79
Appay V, Zaunders JJ, Papagno L, Sutton J, Jaramillo A, Waters A, Easterbrook P, Grey P, Smith D, McMichael AJ, Cooper DA, Rowland-Jones SL, Kelleher AD (2002) Characterization of CD4+ CTLs ex vivo. J Immunol 168:5954–5958
Jellison ER, Kim SK, Welsh RM (2005) Cutting edge: MHC class II-restricted killing in vivo during viral infection. J Immunol 174:614–618
Paludan C, Bickham K, Nikiforow S, Tsang ML, Goodman K, Hanekom WA, Fonteneau JF, Stevanovic S, Münz C (2002) EBNA1 specific CD4+ Th1 cells kill Burkitt's lymphoma cells. J Immunol 169:1593–1603
Nikiforow S, Bottomly K, Miller G, Münz C (2003) Cytolytic CD4+-T-cell clones reactive to EBNA1 inhibit epstein–barr virus-induced B-cell proliferation. J Virol 77:12088–12104
Stevenson PG, Cardin RD, Christensen JP, Doherty PC (1999) Immunological control of a murine gammaherpesvirus independent of CD8+ T cells. J Gen Virol 80:477–483
Sparks-Thissen RL, Braaten DC, Kreher S, Speck SH, Virgin HW (2004) An optimized CD4 T-cell response can control productive and latent gammaherpesvirus infection. J Virol 78:6827–6835
Robertson KA, Usherwood EJ, Nash AA (2001) Regression of a murine gammaherpesvirus 68-positive B-cell lymphoma mediated by CD4 T lymphocytes. J Virol 75:3480–3482
Fu T, Voo KS, Wang RF (2004) Critical role of EBNA1-specific CD4+ T cells in the control of mouse Burkitt lymphoma in vivo. J Clin Invest 114:542–550
Acknowledgements
We thank the National Cancer Institute (R01CA108609), the Arnold and Mabel Beckman Foundation, and the Alexandrine and Alexander Sinsheimer Foundation for supporting our research (to C.M.). D.S. is a recipient of a predoctoral fellowship from the Schering Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Schmid, D., Dengjel, J., Schoor, O. et al. Autophagy in innate and adaptive immunity against intracellular pathogens. J Mol Med 84, 194–202 (2006). https://doi.org/10.1007/s00109-005-0014-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-005-0014-4