Abstract
The antibacterial target, enoyl-acyl carrier protein (ACP) reductase, is a homotetrameric enzyme that catalyzes the last reductive step of fatty acid biosynthesis. In the present paper, Surflex docking and 3D-QSAR studies viz., CoMFA and topomer CoMFA have been carried out on a series (53 compounds) of enoyl ACP reductase inhibitors. An alignment rule for the compounds was defined using the database in SYBYL-X 2.0 package (Tripos Inc., St. Louis, USA, 2012). Models were validated using a data set obtained by dividing the data set into training set and test set. From the CoMFA, topomer CoMFA models, steric and electrostatic maps were generated to analyze the structural features of the datasets to indicate that data are well fitted with high predictive ability and inhibitory potency. The best predictions were obtained with CoMFA model (q 2 = 0.403, \(r^{ 2}_{\text{pred}} = 0. 8 3 2\)) and topomer CoMFA model (q 2 = 0.540, \(r^{ 2}_{\text{pred}} = 0. 8 9 9\)). These models and Surflex-docking studies revealed that the ether linkage, hydroxyl group, and 1,3,4-oxadiazole were significant for binding to the receptor, and some essential features were identified to deduce some interesting SARs. Comparisons of results obtained from both the methods have helped in the understanding of specific activity of some compounds.
Graphical Abstract
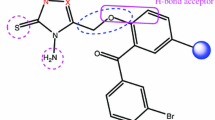
Molecular modeling studies of 1,3,4-oxadiazoles containing substituted phenoxy fragments as enoyl ACP reductase inhibitors Database alignment for enoyl ACP reductase inhibitors.














Similar content being viewed by others
References
Bakhta MA, Yar MS, Abdel-Hamid SG, Al Qasoumi SI, Samad A (2010) Molecular properties prediction, synthesis and antimicrobial activity of some newer oxadiazole derivatives. Eur J Med Chem 45:5862–5869
Cramer RD III (2003) Topomer CoMFA: a design methodology for rapid lead optimization. J Med Chem 46(3):374–388
Cramer RD III, Patterson DE, Bunce JD (1988) Comparative molecular field analysis (CoMFA) 1. Effect of shape on binding of steroids to carrier proteins. J Am Chem Soc 110(18):5959–5967
Cramer RD III, Clark RD, Patterson DE, Ferguson AM (1996) Bioiso-sterism as a molecular diversity descriptor: steric fields of single ‘‘topomeric’’ conformers. J Med Chem 39(16):3060–3069
Gasteiger J, Marsili M (1980) Iterative partial equalization of orbital electronegativity-a rapid access to atomic charges. Tetrahedron 36:3219–3228
Jayatilleke PRN, Nair AC, Zauhar R, Welsh WJ (2000) Computational studies on HIV-1 protease inhibitors: influence of calculated inhibitor-enzyme binding affinities on the statistical quality of 3D-QSAR CoMFA Models. J Med Chem 43:4446–4451
Jilek RJ, Cramer RD III (2004) Topomers: a validated protocol for their self consistent generation. J Chem Inf Comput Sci 44(4):1221–1227
Joshi SD, Nadagouda SG, Khochage A, Pradeepkumar MR, Korat H, Gadaginmath GS (2012a) Synthesis, antibacterial and antifungal activities of 2,5-disubstituted-1,3,4-oxadiazoles. Indian J Heterocycl Chem 22:85–90
Joshi SD, Nadagouda SG, Unale YP, Parkale DB, Ashalata SK, Gadaginamath GS (2012b) Synthesis and evaluation of antibacterial and antifungal activities of some novel 2,5-disubstituted-1,3,4-oxadiazoles. Indian J Heterocycl Chem 21:319–322
Joshi SD, More UA, Pansuriya K, Aminabhavi TM, Gadad AK (2013a) Synthesis and molecular modeling studies of novel pyrrole analogues as antimycobacterial agents. J Saudi Chem Soc. doi:10.1016/j.jscs.2013.09.00
Joshi SD, Nadagouda SG, Pawar S, Gadaginamath GS, Hallikeri CS, Dubey D (2013b) Synthesis and evaluation of antibacterial and antifungal activities of some novel 1,3,4-oxadiazole derivatives. Indian J Heterocycl Chem 22:341–344
Joshi SD, More UA, Aminabhavi TM, Badiger AM (2014a) Two and three-dimensional QSAR studies on a set of antimycobacterial pyrroles: CoMFA, Topomer CoMFA, and HQSAR. Med Chem Res 23:107–126
Joshi SD, More UA, Dixit SR, Korat HH, Aminabhavi TM, Badiger AM (2014b) Synthesis, characterization, biological activity, and 3D-QSAR studies on some novel class of pyrrole derivatives as antitubercular agents. Med Chem Res 23:1123–1147
Karthikeyan MS, Prasad DJ, Mahalingaa M, Holla BS, Kumari NS (2008) Antimicrobial studies of 2,4-dichloro-5-fluorophenyl containing oxadiazoles. Eur J Med Chem 43:25–31
Kollman PA, Cieplak P, Cornell WD, Bayly C (1995) Application of the multi-molecule and multi-conformational RESP methodology to biopolymers: charge derivation for DNA, RNA, and proteins. J Comput Chem 16:1357–1377
McCarthy AD, Hardie DG (1984) Fatty acid synthase: an example of protein evolution by gene fusion. Trends Biochem Sci 9:60–63
McDevitt D, Rosenberg M (2001) Exploiting genomics to discover new antibiotics. Trends Microbiol 9:611–617
Payne DJ, Warren PV, Holmes DJ, Ji Y, Lonsdale JT (2001) Bacterial fatty-acid biosynthesis: a genomics-driven target for antibacterial drug discovery. Drug Discov Today 6:537–544
Srivastava V, Gupta SP, Siddiqi MI, Mishra BN (2010) 3D-QSAR studies on quinazoline antifolate thymidylate synthase inhibitors by CoMFA and CoMSIA models. Eur J Med Chem 45:1560–1571
Tripos International (2012) Sybyl-X 2.0, Tripos International, St. Louis, MO, USA
Vong R, Geladi P, Wold S, Esbensen K (1988) Source contributions to ambient aerosol calculated by discriminate partial least squares regression (PLS). J Chemom 2:281–296
Acknowledgments
The authors gratefully acknowledge the financial support from the Council of Scientific and Industrial Research, New Delhi, India (Letter No. 02(0139)/13/EMR-II dated-12/04/2013). We thank Mr. H. V. Dambal, President, S.E.T’s College of Pharmacy, Dharwad, India, for providing the facilities. The authors are grateful to Mr. Ravi N. Nadiger for his technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Joshi, S.D., Dixit, S.R., More, U.A. et al. 3D-QSAR and molecular docking studies of 1,3,4-oxadiazoles containing substituted phenoxy fragment as inhibitors of enoyl-acyl carrier protein reductase from Escherichia coli . Med Chem Res 23, 4542–4558 (2014). https://doi.org/10.1007/s00044-014-1013-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-014-1013-1